Proteomics Analysis of Aqueous Humor and Rejected Graft in Pig-to-Non-Human Primate Corneal Xenotransplantation
- PMID: 35401527
- PMCID: PMC8986976
- DOI: 10.3389/fimmu.2022.859929
Proteomics Analysis of Aqueous Humor and Rejected Graft in Pig-to-Non-Human Primate Corneal Xenotransplantation
Abstract
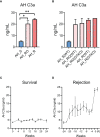
Although pig-to-non-human primate (NHP) corneal xenotransplantation has shown long-term graft survival, xenogeneic antigen-related immune responses are still stronger than allogeneic antigen-associated responses. Therefore, there is an unmet need to investigate major rejection pathways in corneal xenotransplantation, even with immunosuppression. This study aimed to identify biomarkers in aqueous humor for predicting rejection and to investigate rejection-related pathways in grafts from NHPs transplanted with porcine corneas following the administration of steroids combined with tacrolimus/rituximab. NHPs who had received corneas from wild-type (WT) or α-1,3-galactosyltransferase gene-knockout (GTKO) pigs were divided into groups with or without rejection according to clinical examinations. Liquid chromatography-mass spectrometry (LC-MS) was used to analyze the proteomes of corneal tissues or aqueous humor. The biological functions of differentially expressed proteins (DEPs) were assessed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) for pathways and protein-protein interaction network analysis. Among the 66 DEPs in aqueous humor, complement proteins (C3, C5, and C9) and cholesterol metabolic proteins (APOA1 and APOA2) were related to xenogeneic rejection as biomarkers, and alternative pathways of the complement system seemed to be important in xenogeneic graft rejection. Among the 416 DEPs of the cornea, NF-κB1 and proteosomes (PSMD7, PSMA5, and PSMD3) seemed to be related to xenogeneic graft rejection. Additionally, oxidative phosphorylation and leukocyte activation-related pathways are involved in rejection. Overall, our proteomic approach highlights the important role of NF-κB1, proteosomes, oxidative phosphorylation, and leukocyte activation-related inflammation in the cornea and the relevance of complement pathways of the aqueous humor as a predictive biomarker of xenogeneic rejection.
Keywords: aqueous humor; cornea; non-human primate (macaque); pig; proteomics; rejection; xenotransplantation.
Copyright © 2022 Oh, Yoon, Ryu, Kim and Kim.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Initial study of α1,3-galactosyltransferase gene-knockout/CD46 pig full-thickness corneal xenografts in rhesus monkeys.Xenotransplantation. 2017 Jan;24(1). doi: 10.1111/xen.12282. Epub 2017 Jan 5. Xenotransplantation. 2017. PMID: 28054735
-
Efficacy of pig-to-rhesus lamellar corneal xenotransplantation.Invest Ophthalmol Vis Sci. 2011 Aug 22;52(9):6643-50. doi: 10.1167/iovs.11-7273. Invest Ophthalmol Vis Sci. 2011. PMID: 21743020
-
Predictive biomarkers for graft rejection in pig-to-non-human primate corneal xenotransplantation.Xenotransplantation. 2019 Jul;26(4):e12515. doi: 10.1111/xen.12515. Epub 2019 Apr 14. Xenotransplantation. 2019. PMID: 30983050 Free PMC article.
-
Current status of corneal xenotransplantation.Int J Surg. 2015 Nov;23(Pt B):255-260. doi: 10.1016/j.ijsu.2015.07.685. Epub 2015 Jul 29. Int J Surg. 2015. PMID: 26231995 Review.
-
The pathobiology of pig-to-primate xenotransplantation: a historical review.Xenotransplantation. 2016 Mar;23(2):83-105. doi: 10.1111/xen.12219. Epub 2016 Jan 27. Xenotransplantation. 2016. PMID: 26813438 Review.
Cited by
-
Immunology in corneal transplantation-From homeostasis to graft rejection.Transplant Rev (Orlando). 2025 Apr;39(2):100909. doi: 10.1016/j.trre.2025.100909. Epub 2025 Jan 9. Transplant Rev (Orlando). 2025. PMID: 39798206 Review.
-
Xenotransplantation: Current Challenges and Emerging Solutions.Cell Transplant. 2023 Jan-Dec;32:9636897221148771. doi: 10.1177/09636897221148771. Cell Transplant. 2023. PMID: 36644844 Free PMC article. Review.
-
From waste to wealth: Repurposing slaughterhouse waste for xenotransplantation.Front Bioeng Biotechnol. 2023 Feb 3;11:1091554. doi: 10.3389/fbioe.2023.1091554. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 36815880 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

