An Autoantigen Atlas From Human Lung HFL1 Cells Offers Clues to Neurological and Diverse Autoimmune Manifestations of COVID-19
- PMID: 35401574
- PMCID: PMC8987778
- DOI: 10.3389/fimmu.2022.831849
An Autoantigen Atlas From Human Lung HFL1 Cells Offers Clues to Neurological and Diverse Autoimmune Manifestations of COVID-19
Abstract
COVID-19 is accompanied by a myriad of both transient and long-lasting autoimmune responses. Dermatan sulfate (DS), a glycosaminoglycan crucial for wound healing, has unique affinity for autoantigens (autoAgs) from apoptotic cells. DS-autoAg complexes are capable of stimulating autoreactive B cells and autoantibody production. We used DS-affinity proteomics to define the autoantigen-ome of lung fibroblasts and bioinformatics analyses to study the relationship between autoantigenic proteins and COVID-induced alterations. Using DS-affinity, we identified an autoantigen-ome of 408 proteins from human HFL1 cells, at least 231 of which are known autoAgs. Comparing with available COVID data, 352 proteins of the autoantigen-ome have thus far been found to be altered at protein or RNA levels in SARS-CoV-2 infection, 210 of which are known autoAgs. The COVID-altered proteins are significantly associated with RNA metabolism, translation, vesicles and vesicle transport, cell death, supramolecular fibrils, cytoskeleton, extracellular matrix, and interleukin signaling. They offer clues to neurological problems, fibrosis, smooth muscle dysfunction, and thrombosis. In particular, 150 altered proteins are related to the nervous system, including axon, myelin sheath, neuron projection, neuronal cell body, and olfactory bulb. An association with the melanosome is also identified. The findings from our study illustrate a connection between COVID infection and autoimmunity. The vast number of COVID-altered proteins with high intrinsic propensity to become autoAgs offers an explanation for the diverse autoimmune complications in COVID patients. The variety of autoAgs related to mRNA metabolism, translation, and vesicles suggests a need for long-term monitoring of autoimmunity in COVID. The COVID autoantigen atlas we are establishing provides a detailed molecular map for further investigation of autoimmune sequelae of the pandemic, such as "long COVID" syndrome.
Summary sentence: An autoantigen-ome by dermatan sulfate affinity from human lung HFL1 cells may explain neurological and autoimmune manifestations of COVID-19.
Keywords: COVID-19; SARS-CoV-2; autoantibodies; autoantigens; autoimmunity; dermatan sulfate.
Copyright © 2022 Wang, Zhang, Roehrl, Roehrl and Roehrl.
Conflict of interest statement
JW is the founder and Chief Scientific Officer of Curandis. MWR and VR are volunteers of Curandis. MHR is a member of the Scientific Advisory Boards of Trans-Hit Bio (Azenta Life Sciences), Proscia, and Universal DX. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
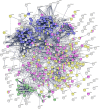
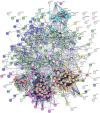
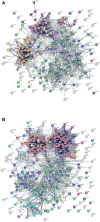

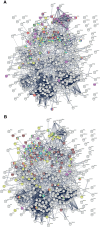
Update of
-
An Autoantigen Atlas from Human Lung HFL1 Cells Offers Clues to Neurological and Diverse Autoimmune Manifestations of COVID-19.bioRxiv [Preprint]. 2021 Jan 24:2021.01.24.427965. doi: 10.1101/2021.01.24.427965. bioRxiv. 2021. Update in: Front Immunol. 2022 Mar 24;13:831849. doi: 10.3389/fimmu.2022.831849. PMID: 33501444 Free PMC article. Updated. Preprint.
References
-
- Luteijn RD, Van Diemen F, Blomen VA, Boer IGJ, Manikam Sadasivam S, van Kuppevelt TH, et al. A Genome-Wide Haploid Genetic Screen Identifies Heparan Sulfate-Associated Genes and the Macropinocytosis Modulator TMED10 as Factors Supporting Vaccinia Virus Infection. J Virol (2019) 93(13):e02160–18. doi: 10.1128/JVI.02160-18 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

