Beta-Amylase and Phosphatidic Acid Involved in Recalcitrant Seed Germination of Chinese Chestnut
- PMID: 35401618
- PMCID: PMC8990265
- DOI: 10.3389/fpls.2022.828270
Beta-Amylase and Phosphatidic Acid Involved in Recalcitrant Seed Germination of Chinese Chestnut
Abstract
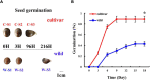
Chinese chestnut (Castanea mollissima), a species with recalcitrant seeds, is an important source of nuts and forest ecosystem services. The germination rate of recalcitrant seeds is low in natural habitats and decreases under conditions of desiccation and low temperature. The germination rate of cultivated Chinese chestnut seeds is significantly higher than that of wild seeds. To explore the reasons for the higher germination rate of cultivated seeds in Chinese chestnut, 113,524 structural variants (SVs) between the wild and cultivated Chinese chestnut genomes were detected through genome comparison. Genotyping these SVs in 60 Chinese chestnut accessions identified allele frequency changes during Chinese chestnut domestication, and some SVs are overlapping genes for controlling seed germination. Transcriptome analysis revealed downregulation of the abscisic acid synthesis genes and upregulation of the beta-amylase synthesis genes in strongly selected genes of cultivated seeds. On the other hand, hormone and enzyme activity assays indicated a decrease in endogenous ABA level and an increase in beta-amylase activity in cultivated seeds. These results shed light on the higher germination rate of cultivated seeds. Moreover, phosphatidic acid synthesis genes are highly expressed in seed germination stages of wild Chinese chestnut and may play a role in recalcitrant seed germination. These findings provide new insight into the regulation of wild seed germination and promote natural regeneration and succession in forest ecosystems.
Keywords: SVs; amylase; phosphatidic acid; recalcitrant seeds; seed germination.
Copyright © 2022 Liu, Zhang, Zheng, Nie, Wang, Yu, Su, Cao, Qin and Xing.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Effects of GABA and Vigabatrin on the Germination of Chinese Chestnut Recalcitrant Seeds and Its Implications for Seed Dormancy and Storage.Plants (Basel). 2020 Apr 3;9(4):449. doi: 10.3390/plants9040449. Plants (Basel). 2020. PMID: 32260136 Free PMC article.
-
Transcriptomic identification and expression of starch and sucrose metabolism genes in the seeds of Chinese chestnut (Castanea mollissima).J Agric Food Chem. 2015 Jan 28;63(3):929-42. doi: 10.1021/jf505247d. Epub 2015 Jan 13. J Agric Food Chem. 2015. PMID: 25537355
-
Melatonin promotes seed germination under salt stress by regulating ABA and GA3 in cotton (Gossypium hirsutum L.).Plant Physiol Biochem. 2021 May;162:506-516. doi: 10.1016/j.plaphy.2021.03.029. Epub 2021 Mar 19. Plant Physiol Biochem. 2021. PMID: 33773227
-
Signatures of Selection in the Genomes of Chinese Chestnut (Castanea mollissima Blume): The Roots of Nut Tree Domestication.Front Plant Sci. 2018 Jun 25;9:810. doi: 10.3389/fpls.2018.00810. eCollection 2018. Front Plant Sci. 2018. PMID: 29988533 Free PMC article.
-
Physiological aspects of seed recalcitrance: a case study on the tree Aesculus hippocastanum.Tree Physiol. 2016 Sep;36(9):1127-50. doi: 10.1093/treephys/tpw037. Epub 2016 Jun 2. Tree Physiol. 2016. PMID: 27259638 Review.
Cited by
-
Comprehensive curation and validation of genomic datasets for chestnut.Sci Data. 2025 May 24;12(1):860. doi: 10.1038/s41597-025-05162-x. Sci Data. 2025. PMID: 40413228 Free PMC article.
References
-
- Ascencio-Ibáñez J. T., Sozzani R., Lee T. J., Chu T. M., Wolfinger R. D., Cella R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148 436–454. 10.1104/pp.108.121038 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

