A Secondary Metabolism Pathway Involved in the Production of a Putative Toxin Is Expressed at Early Stage of Monilinia laxa Infection
- PMID: 35401637
- PMCID: PMC8988988
- DOI: 10.3389/fpls.2022.818483
A Secondary Metabolism Pathway Involved in the Production of a Putative Toxin Is Expressed at Early Stage of Monilinia laxa Infection
Abstract
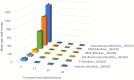
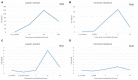
The necrotrophic pathogenic fungus Monilinia laxa causes brown rot disease on stone fruit generating significant yield losses. So far, a limited number of pathogenesis-related virulence factors, such as cell wall degrading enzymes and potential phytotoxins, have been described in Monilinia spp. Using RNA-sequencing data from highly virulent M. laxa ML8L strain at early stages of the infection process (6, 14, 24, and 48 h post-inoculation, hpi) on nectarine and the Pathogen-Host-Interactions (PHI) database, we selected a number of genes for further study and ranked them according to their transcription levels. We identified a class of genes highly expressed at 6 hpi and that their expression decreased to almost undetectable levels at 14 to 48 hpi. Among these genes we found Monilinia__061040 encoding a non-ribosomal peptide synthase (NRPS). Monilinia__061040 together with other five co-regulated genes, forms a secondary metabolism cluster potentially involved in the production of epipolythiodioxopiperazine (ETP) toxin. Quantitative-PCR data confirmed previous RNA sequencing results from the virulent ML8L strain. Interestingly, in a less virulent M. laxa ML5L strain the expression levels of this pathway were reduced compared to the ML8L strain during nectarine infection. In vitro experiments showed that liquid medium containing peach extract mimicked the results observed using nectarines. In fact, upregulation of the NRPS coding gene was also observed in minimal medium suggesting the existence of a fruit-independent mechanism of regulation for this putative toxin biosynthetic pathway that is also downregulated in the less virulent strain. These results emphasize the role of this secondary metabolism pathway during the early stage of brown rot disease development and show alternative models to study the induction of virulence genes in this fungus.
Keywords: NRPS; RNA-seq; brown rot; epipolythiodioxopiperazine; necrotroph; pathogenesis-related pathways; pathogen–host interactions.
Copyright © 2022 Villarino, Rodríguez-Pires, Requena, Melgarejo, De Cal and Espeso.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Balsells-Llauradó M., Silva C. J., Usall J., Vall-llaura N., Serrano-Prieto S., Teixidó N., et al. (2020). Depicting the battle between nectarine and Monilinia laxa: the fruit developmental stage dictates the effectiveness of the host defenses and the pathogen’s infection strategies. Hortic. Res. 7:167. 10.1038/s41438-020-00387-w - DOI - PMC - PubMed

