Eukaryotic mRNA Decapping Activation
- PMID: 35401681
- PMCID: PMC8984151
- DOI: 10.3389/fgene.2022.832547
Eukaryotic mRNA Decapping Activation
Abstract
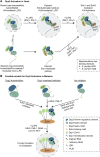
The 5'-terminal cap is a fundamental determinant of eukaryotic gene expression which facilitates cap-dependent translation and protects mRNAs from exonucleolytic degradation. Enzyme-directed hydrolysis of the cap (decapping) decisively affects mRNA expression and turnover, and is a heavily regulated event. Following the identification of the decapping holoenzyme (Dcp1/2) over two decades ago, numerous studies revealed the complexity of decapping regulation across species and cell types. A conserved set of Dcp1/2-associated proteins, implicated in decapping activation and molecular scaffolding, were identified through genetic and molecular interaction studies, and yet their exact mechanisms of action are only emerging. In this review, we discuss the prevailing models on the roles and assembly of decapping co-factors, with considerations of conservation across species and comparison across physiological contexts. We next discuss the functional convergences of decapping machineries with other RNA-protein complexes in cytoplasmic P bodies and compare current views on their impact on mRNA stability and translation. Lastly, we review the current models of decapping activation and highlight important gaps in our current understanding.
Keywords: Cup/Me31B/Tral complex; Dcp1/Dcp2; Edc1; Edc3; Edc4; P bodies; PatL1; mRNA decapping and decay.
Copyright © 2022 Vidya and Duchaine.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

