Population Genetic Structure and Selection Signature Analysis of Beijing Black Pig
- PMID: 35401688
- PMCID: PMC8987279
- DOI: 10.3389/fgene.2022.860669
Population Genetic Structure and Selection Signature Analysis of Beijing Black Pig
Abstract
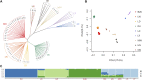
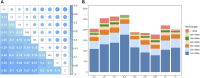
Beijing Black pig is an excellent cultivated black pig breed in China, with desirable body shape, tender meat quality and robust disease resistance. To explore the level of admixture and selection signatures of Beijing Black pigs, a total number of 90 individuals covering nine pig breeds were used in our study, including Beijing Black pig, Large White, Landrace, Duroc, Lantang pig, Luchuan pig, Mashen pig, Huainan pig and Min pig. These animals were resequenced with 18.19 folds mapped read depth on average. Generally, we found that Beijing Black pig was genetically closer to commercial pig breeds by population genetic structure and genetic diversity analysis, and was also affected by Chinese domestic breeds Huainan pig and Min pig. These results are consistent with the cross-breeding history of Beijing Black pig. Selection signal detections were performed on three pig breeds, Beijing Black pig, Duroc and Large White, using three complementary methods (FST, θπ, and XP-EHH). In total, 1,167 significant selected regions and 392 candidate genes were identified. Functional annotations were enriched to pathways related to immune processes and meat and lipid metabolism. Finally, potential candidate genes, influencing meat quality (GPHA2, EHD1, HNF1A, C12orf43, GLTP, TRPV4, MVK, and MMAB), reproduction (PPP2R5B and MAP9), and disease resistance (OASL, ANKRD13A, and GIT2), were further detected by gene annotation analysis. Our results advanced the understanding of the genetic mechanism behind artificial selection of Beijing Black pigs, and provided theoretical basis for the subsequent breeding and genetic research of this breed.
Keywords: beijing black pig; candidate genes; genetic diversity; selection regions; whole-genome sequencing.
Copyright © 2022 Yang, Liu, Zhao, Du, Yu, Wang, Liu, Liu, Jing, Yang, Shi, Zhou and Liu.
Conflict of interest statement
HW, XL, HL, XJ, HY, and GS were employed by Beijing Heiliu Stockbreeding Technology Co.,Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Genome-Wide Re-Sequencing Data Reveals the Population Structure and Selection Signatures of Tunchang Pigs in China.Animals (Basel). 2023 Jun 1;13(11):1835. doi: 10.3390/ani13111835. Animals (Basel). 2023. PMID: 37889708 Free PMC article.
-
Identification of Selection Signatures and Loci Associated with Important Economic Traits in Yunan Black and Huainan Pigs.Genes (Basel). 2023 Mar 5;14(3):655. doi: 10.3390/genes14030655. Genes (Basel). 2023. PMID: 36980926 Free PMC article.
-
Genome-wide detection of selection signatures in Jianli pigs reveals novel cis-regulatory haplotype in EDNRB associated with two-end black coat color.BMC Genomics. 2024 Jan 2;25(1):23. doi: 10.1186/s12864-023-09943-9. BMC Genomics. 2024. PMID: 38166718 Free PMC article.
-
Assessing genomic diversity and signatures of selection in Jiaxian Red cattle using whole-genome sequencing data.BMC Genomics. 2021 Jan 9;22(1):43. doi: 10.1186/s12864-020-07340-0. BMC Genomics. 2021. PMID: 33421990 Free PMC article. Review.
-
Population Structure and Selection Signal Analysis of Nanyang Cattle Based on Whole-Genome Sequencing Data.Genes (Basel). 2024 Mar 11;15(3):351. doi: 10.3390/genes15030351. Genes (Basel). 2024. PMID: 38540410 Free PMC article. Review.
Cited by
-
Genomic Dissection through Whole-Genome Resequencing of Five Local Pig Breeds from Shanghai, China.Animals (Basel). 2023 Dec 1;13(23):3727. doi: 10.3390/ani13233727. Animals (Basel). 2023. PMID: 38067078 Free PMC article.
-
Population genetic analysis based on the polymorphisms mediated by transposons in the genomes of pig.DNA Res. 2024 Apr 1;31(2):dsae008. doi: 10.1093/dnares/dsae008. DNA Res. 2024. PMID: 38447059 Free PMC article.
-
Integrative Analysis of Transcriptomic and Lipidomic Profiles Reveals a Differential Subcutaneous Adipose Tissue Mechanism among Ningxiang Pig and Berkshires, and Their Offspring.Animals (Basel). 2023 Oct 25;13(21):3321. doi: 10.3390/ani13213321. Animals (Basel). 2023. PMID: 37958077 Free PMC article.
-
Genome-Wide Re-Sequencing Data Reveals the Population Structure and Selection Signatures of Tunchang Pigs in China.Animals (Basel). 2023 Jun 1;13(11):1835. doi: 10.3390/ani13111835. Animals (Basel). 2023. PMID: 37889708 Free PMC article.
-
Whole genome and transcriptome analyses identify genetic markers associated with growth traits in Qinchuan black pig.BMC Genomics. 2025 May 12;26(1):469. doi: 10.1186/s12864-025-11627-5. BMC Genomics. 2025. PMID: 40355827 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

