An engineered nano-liposome-human ACE2 decoy neutralizes SARS-CoV-2 Spike protein-induced inflammation in both murine and human macrophages
- PMID: 35401811
- PMCID: PMC8965482
- DOI: 10.7150/thno.66831
An engineered nano-liposome-human ACE2 decoy neutralizes SARS-CoV-2 Spike protein-induced inflammation in both murine and human macrophages
Abstract
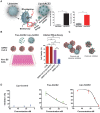
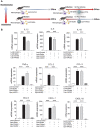
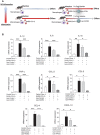
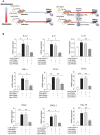
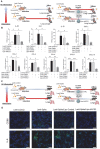
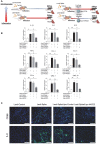
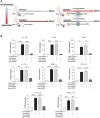
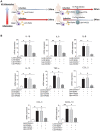
Rationale: Macrophages are the frontline immune cells in response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Angiotensin-converting enzyme 2 (ACE2) serves as the binding receptor to SARS-CoV-2 Spike glycoprotein for fusion and internalization into the human host cells. However, the mechanisms underlying SARS-CoV-2-elicited macrophage inflammatory responses remain elusive. Neutralizing SARS-CoV-2 by human ACE2 (hACE2) decoys has been proposed as a therapeutic approach to ameliorate SARS-CoV-2-stimulated inflammation. This study aims to investigate whether an engineered decoy receptor can abrogate SARS-CoV-2-induced macrophage inflammation. Methods: hACE2 was biotinylated to the surface of nano-liposomes (d = 100 nm) to generate Liposome-human ACE2 complex (Lipo-hACE2). Lentivirus expressing Spike protein (D614G) was also created as a pseudo-SARS-CoV-2 (Lenti-Spike). Liposome-hACE2 was used as a decoy receptor or competitive inhibitor to inhibit SARS-CoV-2 or Lenti-Spike-induced macrophage inflammation in vitro and in vivo. Results: Both SARS-CoV-2 and Lenti-Spike stimulated strong inflammatory responses by inducing the expression of key cytokine and chemokines, including IL-1β, IL-6, TNFα, CCL-2, and CXCL-10, in murine and human macrophages in vitro, whereas Lipo-hACE2 decoy abolished these effects in macrophages. Furthermore, intravenous injection of Lenti-Spike led to increased macrophage and tissue inflammation in wild type mice, which was also abolished by Lipo-hACE2 treatment. Mechanistically, Spike protein stimulated macrophage inflammation by activating canonical NF-κB signaling. RNA sequencing analysis revealed that Lenti-Spike induced over 2,000 differentially expressed genes (DEGs) in murine macrophages, but deficiency of IκB kinase β (IKKβ), a key regulator for NF-κB activation, abrogated Lenti-Spike-elicited macrophage inflammatory responses. Conclusions: We demonstrated that the engineered Lipo-hACE2 acts as a molecular decoy to neutralize SARS-CoV-2 or Spike protein-induced inflammation in both murine and human macrophages, and activation of the canonical IKKβ/NF-κB signaling is essential for SARS-CoV-2-elicited macrophage inflammatory responses.
Keywords: IKKβ/NF-κB signaling; Liposome-Human ACE2; SARS-CoV-2; Spike Protein; myeloid-specific IKKβ knockout.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


Similar articles
-
SARS-CoV-2/ACE2 Interaction Suppresses IRAK-M Expression and Promotes Pro-Inflammatory Cytokine Production in Macrophages.Front Immunol. 2021 Jun 23;12:683800. doi: 10.3389/fimmu.2021.683800. eCollection 2021. Front Immunol. 2021. PMID: 34248968 Free PMC article.
-
Selective Inhibition of the Interaction between SARS-CoV-2 Spike S1 and ACE2 by SPIDAR Peptide Induces Anti-Inflammatory Therapeutic Responses.J Immunol. 2021 Nov 15;207(10):2521-2533. doi: 10.4049/jimmunol.2100144. Epub 2021 Oct 13. J Immunol. 2021. PMID: 34645689 Free PMC article.
-
SARS-CoV-2 deregulates the vascular and immune functions of brain pericytes via Spike protein.Neurobiol Dis. 2021 Dec;161:105561. doi: 10.1016/j.nbd.2021.105561. Epub 2021 Nov 13. Neurobiol Dis. 2021. PMID: 34780863 Free PMC article.
-
The expression of hACE2 receptor protein and its involvement in SARS-CoV-2 entry, pathogenesis, and its application as potential therapeutic target.Tumour Biol. 2021;43(1):177-196. doi: 10.3233/TUB-200084. Tumour Biol. 2021. PMID: 34420993 Review.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
Cited by
-
Paternal hypercholesterolemia elicits sex-specific exacerbation of atherosclerosis in offspring.JCI Insight. 2024 Sep 10;9(17):e179291. doi: 10.1172/jci.insight.179291. JCI Insight. 2024. PMID: 39253968 Free PMC article.
-
Leveraging immunoliposomes as nanocarriers against SARS-CoV-2 and its emerging variants.Asian J Pharm Sci. 2023 Nov;18(6):100855. doi: 10.1016/j.ajps.2023.100855. Epub 2023 Oct 20. Asian J Pharm Sci. 2023. PMID: 38125653 Free PMC article. Review.
-
Paternal phthalate exposure-elicited offspring metabolic disorders are associated with altered sperm small RNAs in mice.Environ Int. 2023 Feb;172:107769. doi: 10.1016/j.envint.2023.107769. Epub 2023 Jan 23. Environ Int. 2023. PMID: 36709676 Free PMC article.
-
Nanoparticles for Coronavirus Control.Nanomaterials (Basel). 2022 May 9;12(9):1602. doi: 10.3390/nano12091602. Nanomaterials (Basel). 2022. PMID: 35564311 Free PMC article. Review.
-
Adipocyte-Derived PXR Signaling Is Dispensable for Diet-Induced Obesity and Metabolic Disorders in Mice.Drug Metab Dispos. 2023 Sep;51(9):1207-1215. doi: 10.1124/dmd.123.001311. Epub 2023 May 25. Drug Metab Dispos. 2023. PMID: 37230767 Free PMC article.
References
-
- Liu J, Liu Y, Xia H, Zou J, Weaver SC, Swanson KA. et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature. 2021;596:273–5. - PubMed
-
- Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F. et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38:970–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

