Inhibitor screening using microarray identifies the high capacity of neutralizing antibodies to Spike variants in SARS-CoV-2 infection and vaccination
- PMID: 35401825
- PMCID: PMC8965487
- DOI: 10.7150/thno.67038
Inhibitor screening using microarray identifies the high capacity of neutralizing antibodies to Spike variants in SARS-CoV-2 infection and vaccination
Abstract
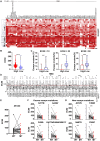
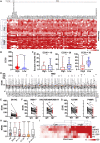
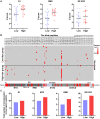
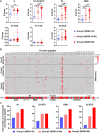
Rationale: Mutations of SARS-CoV-2, which is responsible for coronavirus disease 2019 (COVID-19), could impede drug development and reduce the efficacy of COVID-19 vaccines. Here, we developed a multiplexed Spike-ACE2 Inhibitor Screening (mSAIS) assay that can measure the neutralizing effect of antibodies across numerous variants of the coronavirus's Spike (S) protein simultaneously. Methods: The SARS-CoV-2 spike variant protein microarrays were prepared by printing 72 S variants onto a chemically-modified glass slides. The neutralization potential of purified anti-S antibodies and serum from convalescent COVID-19 patients and vaccinees to S variants were assessed with the mSAIS assay. Results: We identified new S mutations that are sensitive and resistant to neutralization. Serum from both infected and vaccinated groups with a high titer of neutralizing antibodies (NAbs) displayed a broader capacity to neutralize S variants than serum with low titer NAbs. These data were validated using serum from a large vaccinated cohort (n = 104) with a tiled S peptide microarray. In addition, similar results were obtained using a SARS-CoV-2 pseudovirus neutralization assay specific for wild-type S and five prevalent S variants (D614G, B.1.1.7, B.1.351, P.1, B.1.617.2), thus demonstrating that high antibody diversity is associated with high NAb titers. Conclusions: Our results demonstrate the utility of the mSAIS platform in screening NAbs. Moreover, we show that heterogeneous antibody populations provide a more protective effect against S variants, which may help direct COVID-19 vaccine and drug development.
Keywords: SARS-CoV-2; Spike; microarray; mutation; neutralizing antibody.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
Heterogeneous SARS-CoV-2-Neutralizing Activities After Infection and Vaccination.Front Immunol. 2022 May 30;13:888794. doi: 10.3389/fimmu.2022.888794. eCollection 2022. Front Immunol. 2022. PMID: 35711424 Free PMC article.
-
Surface-modified measles vaccines encoding oligomeric, prefusion-stabilized SARS-CoV-2 spike glycoproteins boost neutralizing antibody responses to Omicron and historical variants, independent of measles seropositivity.mBio. 2024 Feb 14;15(2):e0292823. doi: 10.1128/mbio.02928-23. Epub 2024 Jan 9. mBio. 2024. PMID: 38193729 Free PMC article.
-
Comparative analysis of the neutralizing activity against SARS-CoV-2 Wuhan-Hu-1 strain and variants of concern: Performance evaluation of a pseudovirus-based neutralization assay.Front Immunol. 2022 Sep 26;13:981693. doi: 10.3389/fimmu.2022.981693. eCollection 2022. Front Immunol. 2022. PMID: 36225911 Free PMC article.
-
Neutralizing Antibodies Against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variants Induced by Natural Infection or Vaccination: A Systematic Review and Pooled Analysis.Clin Infect Dis. 2022 Mar 1;74(4):734-742. doi: 10.1093/cid/ciab646. Clin Infect Dis. 2022. PMID: 34302458 Free PMC article.
-
SARS-CoV-2 spike S2-specific neutralizing antibodies.Emerg Microbes Infect. 2023 Dec;12(2):2220582. doi: 10.1080/22221751.2023.2220582. Emerg Microbes Infect. 2023. PMID: 37254830 Free PMC article. Review.
Cited by
-
Clinical and humoral immune response characterization of SARS-CoV-2 Omicron BA.2.38 infection in pediatric patients.Heliyon. 2023 Jul 11;9(7):e18093. doi: 10.1016/j.heliyon.2023.e18093. eCollection 2023 Jul. Heliyon. 2023. PMID: 37519697 Free PMC article.
-
A highly sensitive bead-based flow cytometric competitive binding assay to detect SARS-CoV-2 neutralizing antibody activity.Front Immunol. 2022 Nov 30;13:1041860. doi: 10.3389/fimmu.2022.1041860. eCollection 2022. Front Immunol. 2022. PMID: 36532082 Free PMC article.
References
-
- Del Rio C, Malani P. COVID-19 in 2021-continuing uncertainty. JAMA. 2021;325:1389–90. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

