Clinicopathologic and Genomic Features in Triple-Negative Breast Cancer Between Special and No-Special Morphologic Pattern
- PMID: 35402236
- PMCID: PMC8989735
- DOI: 10.3389/fonc.2022.830124
Clinicopathologic and Genomic Features in Triple-Negative Breast Cancer Between Special and No-Special Morphologic Pattern
Abstract
Background: Triple-negative breast cancer (TNBC) is refractory and heterogeneous, comprising various entities with divergent phenotype, biology, and clinical presentation. As an aggressive subtype, Chinese TNBC patients with special morphologic patterns (STs) were restricted to its incidence of 10-15% in total TNBC population.
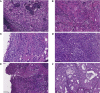
Methods: We recruited 89 patients with TNBC at Guangdong Provincial People's Hospital (GDPH) from October 2014 to May 2021, comprising 72 cases of invasive ductal carcinoma of no-special type (NSTs) and 17 cases of STs. The clinical data of these patients was collected and statistically analyzed. Formalin-fixed, paraffin-embedded (FFPE) tumor tissues and matched blood samples were collected for targeted next-generation sequencing (NGS) with cancer-related, 520- or 33-gene assay. Immunohistochemical analysis of FFPE tissue sections was performed using anti-programmed cell death-ligand 1(PD-L1) and anti-androgen receptor antibodies.
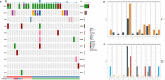
Results: Cases with NSTs presented with higher histologic grade and Ki-67 index rate than ST patients (NSTs to STs: grade I/II/III 1.4%, 16.7%,81.9% vs 0%, 29.4%, 58.8%; p<0.05; Ki-67 ≥30%: 83.3% vs. 58.8%, p<0.05), while androgen receptor (AR) and PD-L1 positive (combined positive score≥10) rates were lower than of STs cases (AR: 11.1% vs. 47.1%; PD-L1: 9.6% vs. 33.3%, p<0.05). The most commonly altered genes were TP53 (88.7%), PIK3CA (26.8%), MYC (18.3%) in NSTs, and TP53 (68.8%), PIK3CA (50%), JAK3 (18.8%), KMT2C (18.8%) in STs respectively. Compared with NSTs, PIK3CA and TP53 mutation frequency showed difference in STs (47.1% vs 19.4%, p=0.039; 64.7% vs 87.5%, p=0.035).
Conclusions: In TNBC patients with STs, decrease in histologic grade and ki-67 index, as well as increase in PD-L1 and AR expression were observed when compared to those with NSTs, suggesting that TNBC patients with STs may better benefit from immune checkpoint inhibitors and/or AR inhibitors. Additionally, lower TP53 and higher PIK3CA mutation rates were also found in STs patients, providing genetic evidence for deciphering at least partly potential mechanism of action.
Keywords: Chinese breast cancer; PD-L1 (22C3); mutation landscape; special type; triple negative breast cancer.
Copyright © 2022 Li, Chen, Lin, Zhang, Lai, Li, Lin, Guo, Xiao, Mok, Ren, Wen, Cao, Lin, Qi, Liu and Liao.
Conflict of interest statement
Authors X-FQ and YL are employed by OrigiMed Co. Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

