Hepatitis C Virus Infection Cycle-Specific MicroRNA Profiling Reveals Stage-Specific miR-4423-3p Targets RIG-I to Facilitate Infection
- PMID: 35402303
- PMCID: PMC8987439
- DOI: 10.3389/fcimb.2022.851917
Hepatitis C Virus Infection Cycle-Specific MicroRNA Profiling Reveals Stage-Specific miR-4423-3p Targets RIG-I to Facilitate Infection
Abstract
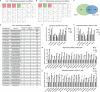
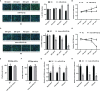
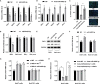
Hepatitis C virus (HCV) infection is one of the main causes of chronic liver diseases, the disorders of which involve multiple pathological processes and elements including host factors such as non-coding small RNAs. Although several genes have been reported to be correlated with HCV infection, the potential regulatory network has not been deciphered clearly. By small RNA sequencing, we clarified the expression profile of microRNAs (miRNAs) in HCV-infected Huh7 and Huh7.5.1 cells and identified 6 dysregulated miRNAs with the same expression trend and 32 dysregulated miRNAs with different expression trends during different stages of HCV life cycle. By looking into each infection stage, we found that 6 miRNAs were entry stage specific, 4 miRNAs were replication stage specific, and 1 miRNA was related to the transmission stage. Moreover, due to the fact that Huh7.5.1 cells have a retinoic acid-inducible gene 1 (RIG-I) mutation which causes reduced production of interferons (IFNs), we here focused on the miRNAs of different trends to decipher the RIG-I/IFN specific miRNAs. Among them, miR-4423-3p showed a significant promotive effect on HCV infection by suppressing RIG-I/IFN pathway through direct binding to RIG-I mRNA. Together, the results displayed novel insights into the miRNA regulatory networks in HCV infection and progression, thus providing a prosperous perspective into the establishment of novel therapeutic and diagnostic targets of the disease.
Keywords: RIG-I; hepatitis C virus; miR-4423-3p; microRNA; small RNA sequencing.
Copyright © 2022 Qian, Wu, Xu and Qi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Bandyopadhyay S., Friedman R. C., Marquez R. T., Keck K., Kong B., Icardi M. S., et al. (2011). Hepatitis C Virus Infection and Hepatic Stellate Cell Activation Downregulate miR-29: miR-29 Overexpression Reduces Hepatitis C Viral Abundance in Culture. J. Infect. Dis. 203 (12), 1753–1762. doi: 10.1093/infdis/jir186 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

