Recruitment of Mobile Genetic Elements for Diverse Cellular Functions in Prokaryotes
- PMID: 35402511
- PMCID: PMC8987985
- DOI: 10.3389/fmolb.2022.821197
Recruitment of Mobile Genetic Elements for Diverse Cellular Functions in Prokaryotes
Abstract
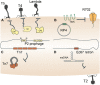
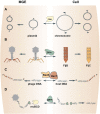
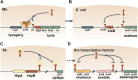
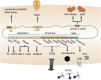
Prokaryotic genomes are replete with mobile genetic elements (MGE) that span a continuum of replication autonomy. On numerous occasions during microbial evolution, diverse MGE lose their autonomy altogether but, rather than being quickly purged from the host genome, assume a new function that benefits the host, rendering the immobilized MGE subject to purifying selection, and resulting in its vertical inheritance. This mini-review highlights the diversity of the repurposed (exapted) MGE as well as the plethora of cellular functions that they perform. The principal contribution of the exaptation of MGE and their components is to the prokaryotic functional systems involved in biological conflicts, and in particular, defense against viruses and other MGE. This evolutionary entanglement between MGE and defense systems appears to stem both from mechanistic similarities and from similar evolutionary predicaments whereby both MGEs and defense systems tend to incur fitness costs to the hosts and thereby evolve mechanisms for survival including horizontal mobility, causing host addiction, and exaptation for functions beneficial to the host. The examples discussed demonstrate that the identity of an MGE, overall mobility and relationship with the host cell (mutualistic, symbiotic, commensal, or parasitic) are all factors that affect exaptation.
Keywords: antivirus defense mechanisms; biological conflict systems; exaptation; horizontal gene transfer; mobile genetic element.
Copyright © 2022 Benler and Koonin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire.Nat Rev Genet. 2020 Feb;21(2):119-131. doi: 10.1038/s41576-019-0172-9. Epub 2019 Oct 14. Nat Rev Genet. 2020. PMID: 31611667 Review.
-
Phylogenomics of Cas4 family nucleases.BMC Evol Biol. 2017 Nov 28;17(1):232. doi: 10.1186/s12862-017-1081-1. BMC Evol Biol. 2017. PMID: 29179671 Free PMC article.
-
Selfish, promiscuous and sometimes useful: how mobile genetic elements drive horizontal gene transfer in microbial populations.Philos Trans R Soc Lond B Biol Sci. 2022 Oct 10;377(1861):20210234. doi: 10.1098/rstb.2021.0234. Epub 2022 Aug 22. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35989606 Free PMC article. Review.
-
Landscape of mobile genetic elements and their antibiotic resistance cargo in prokaryotic genomes.Nucleic Acids Res. 2022 Apr 8;50(6):3155-3168. doi: 10.1093/nar/gkac163. Nucleic Acids Res. 2022. PMID: 35323968 Free PMC article.
-
Why do mobile genetic elements transfer DNA of their hosts?Trends Genet. 2024 Nov;40(11):927-938. doi: 10.1016/j.tig.2024.07.008. Epub 2024 Sep 19. Trends Genet. 2024. PMID: 39304387 Review.
Cited by
-
Search for Origins of Anti-CRISPR Proteins by Structure Comparison.CRISPR J. 2023 Jun;6(3):222-231. doi: 10.1089/crispr.2023.0011. CRISPR J. 2023. PMID: 37272863 Free PMC article.
-
Marine viral particles reveal an expansive repertoire of phage-parasitizing mobile elements.Proc Natl Acad Sci U S A. 2022 Oct 25;119(43):e2212722119. doi: 10.1073/pnas.2212722119. Epub 2022 Oct 18. Proc Natl Acad Sci U S A. 2022. PMID: 36256808 Free PMC article.
-
Structural basis of antiphage defense by an ATPase-associated reverse transcriptase.bioRxiv [Preprint]. 2025 Mar 26:2025.03.26.645336. doi: 10.1101/2025.03.26.645336. bioRxiv. 2025. PMID: 40196496 Free PMC article. Preprint.
-
Transposon-encoded nucleases use guide RNAs to selfishly bias their inheritance.bioRxiv [Preprint]. 2023 Mar 29:2023.03.14.532601. doi: 10.1101/2023.03.14.532601. bioRxiv. 2023. Update in: Nature. 2023 Oct;622(7984):863-871. doi: 10.1038/s41586-023-06597-1. PMID: 36993599 Free PMC article. Updated. Preprint.
-
Genome-centric metagenomes unveiling the hidden resistome in an anchialine cave.Environ Microbiome. 2024 Sep 9;19(1):67. doi: 10.1186/s40793-024-00612-2. Environ Microbiome. 2024. PMID: 39252078 Free PMC article.
References
-
- Abe K., Takamatsu T., Sato T. (2017). Mechanism of Bacterial Gene Rearrangement: SprA-Catalyzed Precise DNA Recombination and its Directionality Control by SprB Ensure the Gene Rearrangement and Stable Expression of spsM during Sporulation in Bacillus Subtilis. Nucleic Acids Res. 45, 6669–6683. 10.1093/nar/gkx466 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

