Empowering Data Sharing and Analytics through the Open Data Commons for Traumatic Brain Injury Research
- PMID: 35403104
- PMCID: PMC8985540
- DOI: 10.1089/neur.2021.0061
Empowering Data Sharing and Analytics through the Open Data Commons for Traumatic Brain Injury Research
Abstract
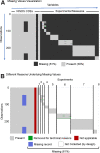
Traumatic brain injury (TBI) is a major public health problem. Despite considerable research deciphering injury pathophysiology, precision therapies remain elusive. Here, we present large-scale data sharing and machine intelligence approaches to leverage TBI complexity. The Open Data Commons for TBI (ODC-TBI) is a community-centered repository emphasizing Findable, Accessible, Interoperable, and Reusable data sharing and publication with persistent identifiers. Importantly, the ODC-TBI implements data sharing of individual subject data, enabling pooling for high-sample-size, feature-rich data sets for machine learning analytics. We demonstrate pooled ODC-TBI data analyses, starting with descriptive analytics of subject-level data from 11 previously published articles (N = 1250 subjects) representing six distinct pre-clinical TBI models. Second, we perform unsupervised machine learning on multi-cohort data to identify persistent inflammatory patterns across different studies, improving experimental sensitivity for pro- versus anti-inflammation effects. As funders and journals increasingly mandate open data practices, ODC-TBI will create new scientific opportunities for researchers and facilitate multi-data-set, multi-dimensional analytics toward effective translation.
Keywords: FAIR principles; Open Data Commons; data sharing; multi-variate analysis; principal component analysis; traumatic brain Injury.
© Austin Chou et al., 2022; Published by Mary Ann Liebert, Inc.
Conflict of interest statement
No competing financial interests exist.
Figures






References
-
- Centers for Disease Control and Prevention. (2015). Traumatic Brain Injury In the United States: Epidemiology and Rehabilitation. National Center for Injury Prevention and Control; Division of Unintentional Injury Prevention: Atlanta, GA.
-
- Dewan, M.C., Rattani, A., Gupta, S., Baticulon, R.E., Hung, Y.-C., Punchak, M., Agrawal, A., Adeleye, A.O., Shrime, M.G., Rubiano, A.M., Rosenfeld, J.V., and Park, K.B. (2018). Estimating the global incidence of traumatic brain injury. J. Neurosurg. 130, 1080–1097. - PubMed
-
- Zaloshnja, E., Miller, T., Langlois, J.A., and Selassie, A.W. (2008). Prevalence of long-term disability from traumatic brain injury in the civilian population of the United States, 2005. J. Head Trauma Rehabil. 23, 394–400. - PubMed
-
- Masel, B.E., and DeWitt, D.S. (2010). Traumatic brain injury: a disease process, not an event. J. Neurotrauma 27, 1529–1540. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
