Phylogenetically diverse Bradyrhizobium genospecies nodulate Bambara groundnut (Vigna subterranea L. Verdc) and soybean (Glycine max L. Merril) in the northern savanna zones of Ghana
- PMID: 35404419
- PMCID: PMC9329091
- DOI: 10.1093/femsec/fiac043
Phylogenetically diverse Bradyrhizobium genospecies nodulate Bambara groundnut (Vigna subterranea L. Verdc) and soybean (Glycine max L. Merril) in the northern savanna zones of Ghana
Abstract
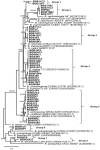
A total of 102 bacterial strains isolated from nodules of three Bambara groundnut and one soybean cultivars grown in nineteen soil samples collected from northern Ghana were characterized using multilocus gene sequence analysis. Based on a concatenated sequence analysis (glnII-rpoB-recA-gyrB-atpD-dnaK), 54 representative strains were distributed in 12 distinct lineages, many of which were placed mainly in the Bradyrhizobium japonicum and Bradyrhizobium elkanii supergroups. Twenty-four of the 54 representative strains belonged to seven putative novel species, while 30 were conspecific with four recognized Bradyrhizobium species. The nodA phylogeny placed all the representative strains in the cosmopolitan nodA clade III. The strains were further separated in seven nodA subclusters with reference strains mainly of African origin. The nifH phylogeny was somewhat congruent with the nodA phylogeny, but both symbiotic genes were mostly incongruent with the core housekeeping gene phylogeny indicating that the strains acquired their symbiotic genes horizontally from distantly related Bradyrhizobium species. Using redundancy analysis, the distribution of genospecies was found to be influenced by the edaphic factors of the respective sampling sites. In general, these results mainly underscore the high genetic diversity of Bambara groundnut-nodulating bradyrhizobia in Ghanaian soils and suggest a possible vast resource of adapted inoculant strains.
Keywords: Bradyrhizobium; Bambara groundnut; biological nitrogen fixation; housekeeping genes; multi-locus sequence analysis; soybean.
© The Author(s) 2022. Published by Oxford University Press on behalf of FEMS.
Figures





References
-
- Abaidoo RC, Keyser HH, Singleton PWet al. Bradyrhizobium spp. (TGx) isolates nodulating the new soybean cultivars in Africa are diverse and distinct from bradyrhizobia that nodulate North American soybeans. Int J Syst Evol Microbiol. 2000;50:225–34. - PubMed
-
- Abu HB, Buah SSJ.. Characterization of Bambara groundnut landraces and their evaluation by farmers in the upper west region of Ghana. J Dev Sustain Agric. 2011;6:64–74.
-
- Aidoo R, Osei-Mensah J, Abaidoo RCet al. Factors influencing soyabean production and willingness to pay for inoculum use in Northern Ghana. J Exp Agric Int. 2013;290–301.
-
- Alhassan GA, Egbe MO.. Participatory rural appraisal of Bambara groundnut (Vigna subterranea (L.) Verdc.) production in Southern Guinea Savanna of Nigeria. Agric Sci. 2013;1:18–31.
-
- Altschul SF, Gish W, Miller Wet al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. - PubMed

