A spatially aware likelihood test to detect sweeps from haplotype distributions
- PMID: 35404934
- PMCID: PMC9022890
- DOI: 10.1371/journal.pgen.1010134
A spatially aware likelihood test to detect sweeps from haplotype distributions
Abstract
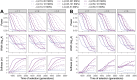
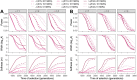
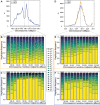
The inference of positive selection in genomes is a problem of great interest in evolutionary genomics. By identifying putative regions of the genome that contain adaptive mutations, we are able to learn about the biology of organisms and their evolutionary history. Here we introduce a composite likelihood method that identifies recently completed or ongoing positive selection by searching for extreme distortions in the spatial distribution of the haplotype frequency spectrum along the genome relative to the genome-wide expectation taken as neutrality. Furthermore, the method simultaneously infers two parameters of the sweep: the number of sweeping haplotypes and the "width" of the sweep, which is related to the strength and timing of selection. We demonstrate that this method outperforms the leading haplotype-based selection statistics, though strong signals in low-recombination regions merit extra scrutiny. As a positive control, we apply it to two well-studied human populations from the 1000 Genomes Project and examine haplotype frequency spectrum patterns at the LCT and MHC loci. We also apply it to a data set of brown rats sampled in NYC and identify genes related to olfactory perception. To facilitate use of this method, we have implemented it in user-friendly open source software.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

