Extract, transform, load framework for the conversion of health databases to OMOP
- PMID: 35404974
- PMCID: PMC9000122
- DOI: 10.1371/journal.pone.0266911
Extract, transform, load framework for the conversion of health databases to OMOP
Abstract
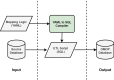
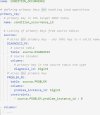
Common data models standardize the structures and semantics of health datasets, enabling reproducibility and large-scale studies that leverage the data from multiple locations and settings. The Observational Medical Outcomes Partnership Common Data Model (OMOP CDM) is one of the leading common data models. While there is a strong incentive to convert datasets to OMOP, the conversion is time and resource-intensive, leaving the research community in need of tools for mapping data to OMOP. We propose an extract, transform, load (ETL) framework that is metadata-driven and generic across source datasets. The ETL framework uses a new data manipulation language (DML) that organizes SQL snippets in YAML. Our framework includes a compiler that converts YAML files with mapping logic into an ETL script. Access to the ETL framework is available via a web application, allowing users to upload and edit YAML files via web editor and obtain an ETL SQL script for use in development environments. The structure of the DML maximizes readability, refactoring, and maintainability, while minimizing technical debt and standardizing the writing of ETL operations for mapping to OMOP. Our framework also supports transparency of the mapping process and reuse by different institutions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

