Radiomics-Based Method for Predicting the Glioma Subtype as Defined by Tumor Grade, IDH Mutation, and 1p/19q Codeletion
- PMID: 35406550
- PMCID: PMC8997070
- DOI: 10.3390/cancers14071778
Radiomics-Based Method for Predicting the Glioma Subtype as Defined by Tumor Grade, IDH Mutation, and 1p/19q Codeletion
Abstract
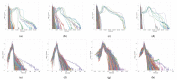
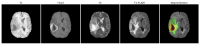
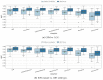
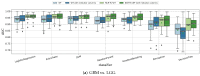
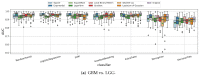
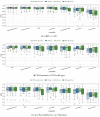
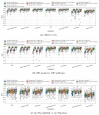
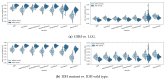
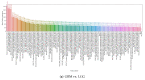
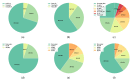
Gliomas are among the most common types of central nervous system (CNS) tumors. A prompt diagnosis of the glioma subtype is crucial to estimate the prognosis and personalize the treatment strategy. The objective of this study was to develop a radiomics pipeline based on the clinical Magnetic Resonance Imaging (MRI) scans to noninvasively predict the glioma subtype, as defined based on the tumor grade, isocitrate dehydrogenase (IDH) mutation status, and 1p/19q codeletion status. A total of 212 patients from the public retrospective The Cancer Genome Atlas Low Grade Glioma (TCGA-LGG) and The Cancer Genome Atlas Glioblastoma Multiforme (TCGA-GBM) datasets were used for the experiments and analyses. Different settings in the radiomics pipeline were investigated to improve the classification, including the Z-score normalization, the feature extraction strategy, the image filter applied to the MRI images, the introduction of clinical information, ComBat harmonization, the classifier chain strategy, etc. Based on numerous experiments, we finally reached an optimal pipeline for classifying the glioma tumors. We then tested this final radiomics pipeline on the hold-out test data with 51 randomly sampled random seeds for reliable and robust conclusions. The results showed that, after tuning the radiomics pipeline, the mean AUC improved from 0.8935 (±0.0351) to 0.9319 (±0.0386), from 0.8676 (±0.0421) to 0.9283 (±0.0333), and from 0.6473 (±0.1074) to 0.8196 (±0.0702) in the test data for predicting the tumor grade, IDH mutation, and 1p/19q codeletion status, respectively. The mean accuracy for predicting the five glioma subtypes also improved from 0.5772 (±0.0816) to 0.6716 (±0.0655). Finally, we analyzed the characteristics of the radiomic features that best distinguished the glioma grade, the IDH mutation, and the 1p/19q codeletion status, respectively. Apart from the promising prediction of the glioma subtype, this study also provides a better understanding of the radiomics model development and interpretability. The results in this paper are replicable with our python codes publicly available in github.
Keywords: 1p/19q codeletion; IDH mutation; glioblastomas; gliomas; radiomics; tumor grade.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures











References
-
- Louis D.N., Perry A., Reifenberger G., Von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. - DOI - PubMed
-
- Qi S., Yu L., Gui S., Ding Y., Han H., Zhang X., Wu L., Yao F. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012;103:269–273. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

