NGLY1 Deficiency, a Congenital Disorder of Deglycosylation: From Disease Gene Function to Pathophysiology
- PMID: 35406718
- PMCID: PMC8997433
- DOI: 10.3390/cells11071155
NGLY1 Deficiency, a Congenital Disorder of Deglycosylation: From Disease Gene Function to Pathophysiology
Abstract
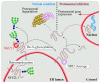
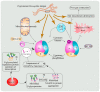
N-Glycanase 1 (NGLY1) is a cytosolic enzyme involved in removing N-linked glycans of misfolded N-glycoproteins and is considered to be a component of endoplasmic reticulum-associated degradation (ERAD). The 2012 identification of recessive NGLY1 mutations in a rare multisystem disorder has led to intense research efforts on the roles of NGLY1 in animal development and physiology, as well as the pathophysiology of NGLY1 deficiency. Here, we present a review of the NGLY1-deficient patient phenotypes, along with insights into the function of this gene from studies in rodent and invertebrate animal models, as well as cell culture and biochemical experiments. We will discuss critical processes affected by the loss of NGLY1, including proteasome bounce-back response, mitochondrial function and homeostasis, and bone morphogenetic protein (BMP) signaling. We will also cover the biologically relevant targets of NGLY1 and the genetic modifiers of NGLY1 deficiency phenotypes in animal models. Together, these discoveries and disease models have provided a number of avenues for preclinical testing of potential therapeutic approaches for this disease.
Keywords: AMPK signaling; BMP signaling; ER-associated degradation (ERAD); N-glycosylation; congenital disorder of deglycosylation (CDDG); deglycosylation; human developmental disorder; mitochondrial abnormality; proteasome; rare disease.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study, in the writing of the manuscript, or in the decision to publish the manuscript.
Figures

References
-
- Stanley P., Taniguchi N., Aebi M. N-Glycans. In: Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., Prestegard J.H., et al., editors. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2015. pp. 99–111. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

