Biochemistry and Molecular Basis of Intracellular Flavonoid Transport in Plants
- PMID: 35406945
- PMCID: PMC9002769
- DOI: 10.3390/plants11070963
Biochemistry and Molecular Basis of Intracellular Flavonoid Transport in Plants
Abstract
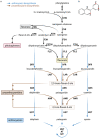
Flavonoids are a biochemically diverse group of specialized metabolites in plants that are derived from phenylalanine. While the biosynthesis of the flavonoid aglycone is highly conserved across species and well characterized, numerous species-specific decoration steps and their relevance remained largely unexplored. The flavonoid biosynthesis takes place at the cytosolic side of the endoplasmatic reticulum (ER), but accumulation of various flavonoids was observed in the central vacuole. A universal explanation for the subcellular transport of flavonoids has eluded researchers for decades. Current knowledge suggests that a glutathione S-transferase-like protein (ligandin) protects anthocyanins and potentially proanthocyanidin precursors during the transport to the central vacuole. ABCC transporters and to a lower extend MATE transporters sequester anthocyanins into the vacuole. Glycosides of specific proanthocyanidin precursors are sequestered through MATE transporters. A P-ATPase in the tonoplast and potentially other proteins generate the proton gradient that is required for the MATE-mediated antiport. Vesicle-mediated transport of flavonoids from the ER to the vacuole is considered as an alternative or additional route.
Keywords: ABCC; MATE; anthocyanins; flavones; flavonoid accumulation; flavonoid biosynthesis; flavonoid transport; flavonols; ligandin; proanthocyanidins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Afendi F.M., Okada T., Yamazaki M., Hirai-Morita A., Nakamura Y., Nakamura K., Ikeda S., Takahashi H., Altaf-Ul-Amin M., Darusman L.K., et al. KNApSAcK Family Databases: Integrated Metabolite-Plant Species Databases for Multifaceted Plant Research. Plant Cell Physiol. 2012;53:e1. doi: 10.1093/pcp/pcr165. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

