A Comparison of Network-Based Methods for Drug Repurposing along with an Application to Human Complex Diseases
- PMID: 35409062
- PMCID: PMC8999012
- DOI: 10.3390/ijms23073703
A Comparison of Network-Based Methods for Drug Repurposing along with an Application to Human Complex Diseases
Abstract
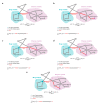
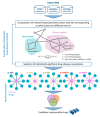
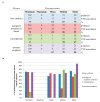
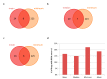
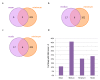
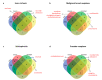
Drug repurposing strategy, proposing a therapeutic switching of already approved drugs with known medical indications to new therapeutic purposes, has been considered as an efficient approach to unveil novel drug candidates with new pharmacological activities, significantly reducing the cost and shortening the time of de novo drug discovery. Meaningful computational approaches for drug repurposing exploit the principles of the emerging field of Network Medicine, according to which human diseases can be interpreted as local perturbations of the human interactome network, where the molecular determinants of each disease (disease genes) are not randomly scattered, but co-localized in highly interconnected subnetworks (disease modules), whose perturbation is linked to the pathophenotype manifestation. By interpreting drug effects as local perturbations of the interactome, for a drug to be on-target effective against a specific disease or to cause off-target adverse effects, its targets should be in the nearby of disease-associated genes. Here, we used the network-based proximity measure to compute the distance between the drug module and the disease module in the human interactome by exploiting five different metrics (minimum, maximum, mean, median, mode), with the aim to compare different frameworks for highlighting putative repurposable drugs to treat complex human diseases, including malignant breast and prostate neoplasms, schizophrenia, and liver cirrhosis. Whilst the standard metric (that is the minimum) for the network-based proximity remained a valid tool for efficiently screening off-label drugs, we observed that the other implemented metrics specifically predicted further interesting drug candidates worthy of investigation for yielding a potentially significant clinical benefit.
Keywords: drug repurposing; network medicine; network theory.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Caldera M., Buphamalai P., Müller F., Menche J. Interactome-based approaches to human disease. Curr. Opin. Syst. Biol. 2017;3:88–94. doi: 10.1016/j.coisb.2017.04.015. - DOI
-
- Silverman E.K., Schmidt H.H.H.W., Anastasiadou E., Altucci L., Angelini M., Basimon L., Balligand J.-L., Benincasa G., Capasso G., Conte F., et al. Molecular networks in Network Medicine: Development and applications. WIREs Syst. Biol. Med. 2020;12:e1489. doi: 10.1002/wsbm.1489. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

