Pyruvate Dehydrogenase Kinase Inhibition by Dichloroacetate in Melanoma Cells Unveils Metabolic Vulnerabilities
- PMID: 35409102
- PMCID: PMC8999016
- DOI: 10.3390/ijms23073745
Pyruvate Dehydrogenase Kinase Inhibition by Dichloroacetate in Melanoma Cells Unveils Metabolic Vulnerabilities
Abstract
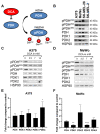
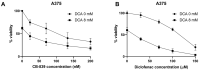
Melanoma is characterized by high glucose uptake, partially mediated through elevated pyruvate dehydrogenase kinase (PDK), making PDK a potential treatment target in melanoma. We aimed to reduce glucose uptake in melanoma cell lines through PDK inhibitors dichloroacetate (DCA) and AZD7545 and through PDK knockdown, to inhibit cell growth and potentially unveil metabolic co-vulnerabilities resulting from PDK inhibition. MeWo cells were most sensitive to DCA, while SK-MEL-2 was the least sensitive, with IC50 values ranging from 13.3 to 27.0 mM. DCA strongly reduced PDH phosphorylation and increased the oxygen consumption rate:extracellular acidification rate (OCR:ECAR) ratio up to 6-fold. Knockdown of single PDK isoforms had similar effects on PDH phosphorylation and OCR:ECAR ratio as DCA but did not influence sensitivity to DCA. Growth inhibition by DCA was synergistic with the glutaminase inhibitor CB-839 (2- to 5-fold sensitization) and with diclofenac, known to inhibit monocarboxylate transporters (MCTs) (3- to 8-fold sensitization). CB-839 did not affect the OCR:ECAR response to DCA, whereas diclofenac strongly inhibited ECAR and further increased the OCR:ECAR ratio. We conclude that in melanoma cell lines, DCA reduces proliferation through reprogramming of cellular metabolism and synergizes with other metabolically targeted drugs.
Keywords: dichloroacetate; melanoma; metabolic reprogramming; metabolism.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
-
- Robert C., Grob J.J., Stroyakovskiy D., Karaszewska B., Hauschild A., Levchenko E., Chiarion Sileni V., Schachter J., Garbe C., Bondarenko I., et al. Five-Year Outcomes with Dabrafenib plus Trametinib in Metastatic Melanoma. N. Engl. J. Med. 2019;381:626–636. doi: 10.1056/NEJMoa1904059. - DOI - PubMed
-
- Shi H., Hugo W., Kong X., Hong A., Koya R.C., Moriceau G., Chodon T., Guo R., Johnson D.B., Dahlman K.B., et al. Acquired Resistance and Clonal Evolution in Melanoma during BRAF Inhibitor Therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

