BRCA1/2 Serves as a Biomarker for Poor Prognosis in Breast Carcinoma
- PMID: 35409110
- PMCID: PMC8998777
- DOI: 10.3390/ijms23073754
BRCA1/2 Serves as a Biomarker for Poor Prognosis in Breast Carcinoma
Abstract
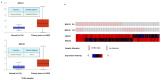
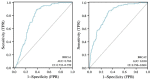
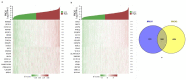
BRCA1/2 are breast cancer susceptibility genes that are involved in DNA repair and transcriptional control. They are dysregulated in breast cancer, making them attractive therapeutic targets. Here, we performed a systematic multiomics analysis to expound BRCA1/2 functions as prognostic biomarkers in breast cancer. First, using different web-based bioinformatics platforms (Oncomine, TIMER 2.0, UALCAN, and cBioportal), the expression of BRCA1/2 was assessed. Then, the R package was used to analyze the diagnostic value of BRCA1/2 in patients. Next, we determined the relationship between BRCA1/2 mRNA expression and prognosis in patients (PrognoScan Database, R2: Kaplan Meier Scanner and Kaplan−Meier Plotter). Subsequently, the association of BRCA1/2 with mutation frequency alteration and copy number alterations in breast cancer was investigated using the cBioportal platform. After that, we identified known and predicted structural genes and proteins essential for BRCA1/2 functions using GeneMania and STRING db. Finally, GO and KEGG pathway enrichment analyses were performed to elucidate the potential biological functions of the co-expression genes of BRCA1/2. The BRCA1/2 mRNA level in breast cancer tissues was considerably higher than in normal tissues, with AUCs of 0.766 and 0.829, respectively. Overexpression of BRCA1/2 was significantly related to the worse overall survival (p < 0.001) and was correlated to clinicopathological characteristics including lymph nodes, estrogen receptors, and progesterone receptors (p < 0.01). The alteration frequencies of both the gens have been checked, and the results show that BRCA1 and BRCA2 show different alteration frequencies. Their mutation sites differ from each other. GO and KEGG showed that BRCA1/2 was mainly enriched in catalytic activity, acting on DNA, chromosomal region, organelle fission, cell cycle, etc. The 20 most frequently changed genes were closely related to BRCA1/2, including PALB2 and RAD51 relatively. Our study provides suggestive evidence of the prognostic role of BRCA1/2 in breast cancer and the therapeutic target for breast cancer. Furthermore, BRCA1/2 may influence BRCA prognosis through catalytic activity, acting on DNA, chromosomal regions, organelle fission, and the cell cycle. Nevertheless, further validation is warranted.
Keywords: BRCA1/2; breast cancer; multiomics; prognostic biomarkers.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

