SSGraphCPI: A Novel Model for Predicting Compound-Protein Interactions Based on Deep Learning
- PMID: 35409140
- PMCID: PMC8998983
- DOI: 10.3390/ijms23073780
SSGraphCPI: A Novel Model for Predicting Compound-Protein Interactions Based on Deep Learning
Abstract
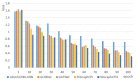
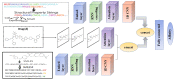
Identifying compound-protein (drug-target, DTI) interactions (CPI) accurately is a key step in drug discovery. Including virtual screening and drug reuse, it can significantly reduce the time it takes to identify drug candidates and provide patients with timely and effective treatment. Recently, more and more researchers have developed CPI's deep learning model, including feature representation of a 2D molecular graph of a compound using a graph convolutional neural network, but this method loses much important information about the compound. In this paper, we propose a novel three-channel deep learning framework, named SSGraphCPI, for CPI prediction, which is composed of recurrent neural networks with an attentional mechanism and graph convolutional neural network. In our model, the characteristics of compounds are extracted from 1D SMILES string and 2D molecular graph. Using both the 1D SMILES string sequence and the 2D molecular graph can provide both sequential and structural features for CPI predictions. Additionally, we select the 1D CNN module to learn the hidden data patterns in the sequence to mine deeper information. Our model is much more suitable for collecting more effective information of compounds. Experimental results show that our method achieves significant performances with RMSE (Root Mean Square Error) = 2.24 and R2 (degree of linear fitting of the model) = 0.039 on the GPCR (G Protein-Coupled Receptors) dataset, and with RMSE = 2.64 and R2 = 0.018 on the GPCR dataset RMSE, which preforms better than some classical deep learning models, including RNN/GCNN-CNN, GCNNet and GATNet.
Keywords: IC50 value; compound properties; compound-protein interactions; deep learning; protein preperties.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Scannell J.W., Blanckley A., Boldon H., Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 2012;11:191–200. - PubMed
-
- Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–36. doi: 10.1021/ci00057a005. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

