The Characterization of the Phloem Protein 2 Gene Family Associated with Resistance to Sclerotinia sclerotiorum in Brassica napus
- PMID: 35409295
- PMCID: PMC8999561
- DOI: 10.3390/ijms23073934
The Characterization of the Phloem Protein 2 Gene Family Associated with Resistance to Sclerotinia sclerotiorum in Brassica napus
Abstract
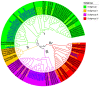
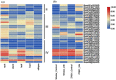
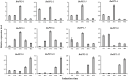
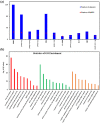
In plants, phloem is not only a vital structure that is used for nutrient transportation, but it is also the location of a response that defends against various stresses, named phloem-based defense (PBD). Phloem proteins (PP2s) are among the predominant proteins in phloem, indicating their potential functional role in PBD. Sclerotinia disease (SD), which is caused by the necrotrophic fungal pathogen S. sclerotiorum (Sclerotinia sclerotiorum), is a devastating disease that affects oil crops, especially Brassica napus (B. napus), mainly by blocking nutrition and water transportation through xylem and phloem. Presently, the role of PP2s in SD resistance is still largely estimated. Therefore, in this study, we identified 62 members of the PP2 gene family in the B. napus genome with an uneven distribution across the 19 chromosomes. A phylogenetic analysis classified the BnPP2s into four clusters (I-IV), with cluster I containing the most members (28 genes) as a consequence of its frequent genome segmental duplication. A comparison of the gene structures and conserved motifs suggested that BnPP2 genes were well conserved in clusters II to IV, but were variable in cluster I. Interestingly, the motifs in different clusters displayed unique features, such as motif 6 specifically existing in cluster III and motif 1 being excluded from cluster IV. These results indicated the possible functional specification of BnPP2s. A transcriptome data analysis showed that the genes in clusters II to IV exhibited dynamic expression alternation in tissues and the stimulation of S. sclerotiorum, suggesting that they could participate in SD resistance. A GWAS analysis of a rapeseed population comprising 324 accessions identified four BnPP2 genes that were potentially responsible for SD resistance and a transgenic study that was conducted by transiently expressing BnPP2-6 in tobacco (Nicotiana tabacum) leaves validated their positive role in regulating SD resistance in terms of reduced lesion size after inoculation with S. sclerotiorum hyphal plugs. This study provides useful information on PP2 gene functions in B. napus and could aid elaborated functional studies on the PP2 gene family.
Keywords: Brassica napus; Sclerotinia disease resistance; phloem protein 2; phylogenetic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Jung J.H., Park C.M. Vascular development in plants: Specification of xylem and phloem tissues. J. Plant Biol. 2007;50:301–305. doi: 10.1007/BF03030658. - DOI
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

