Antisense Oligonucleotide Induction of the hnRNPA1b Isoform Affects Pre-mRNA Splicing of SMN2 in SMA Type I Fibroblasts
- PMID: 35409296
- PMCID: PMC8999010
- DOI: 10.3390/ijms23073937
Antisense Oligonucleotide Induction of the hnRNPA1b Isoform Affects Pre-mRNA Splicing of SMN2 in SMA Type I Fibroblasts
Abstract
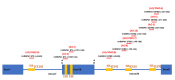
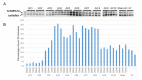
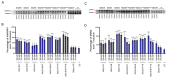
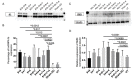
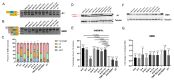
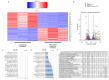
Spinal muscular atrophy (SMA) is a severe, debilitating neuromuscular condition characterised by loss of motor neurons and progressive muscle wasting. SMA is caused by a loss of expression of SMN1 that encodes the survival motor neuron (SMN) protein necessary for the survival of motor neurons. Restoration of SMN expression through increased inclusion of SMN2 exon 7 is known to ameliorate symptoms in SMA patients. As a consequence, regulation of pre-mRNA splicing of SMN2 could provide a potential molecular therapy for SMA. In this study, we explored if splice switching antisense oligonucleotides could redirect the splicing repressor hnRNPA1 to the hnRNPA1b isoform and restore SMN expression in fibroblasts from a type I SMA patient. Antisense oligonucleotides (AOs) were designed to promote exon 7b retention in the mature mRNA and induce the hnRNPA1b isoform. RT-PCR and western blot analysis were used to assess and monitor the efficiency of different AO combinations. A combination of AOs targeting multiple silencing motifs in hnRNPA1 pre-mRNA led to robust hnRNPA1b induction, which, in turn, significantly increased expression of full-length SMN (FL-SMN) protein. A combination of PMOs targeting the same motifs also strongly induced hnRNPA1b isoform, but surprisingly SMN2 exon 5 skipping was detected, and the PMO cocktail did not lead to a significant increase in expression of FL-SMN protein. We further performed RNA sequencing to assess the genome-wide effects of hnRNPA1b induction. Some 3244 genes were differentially expressed between the hnRNPA1b-induced and untreated SMA fibroblasts, which are functionally enriched in cell cycle and chromosome segregation processes. RT-PCR analysis demonstrated that expression of the master regulator of these enrichment pathways, MYBL2 and FOXM1B, were reduced in response to PMO treatment. These findings suggested that induction of hnRNPA1b can promote SMN protein expression, but not at sufficient levels to be clinically relevant.
Keywords: antisense oligonucleotide; hnRNPA1; phosphorodiamidate morpholino oligomer; spinal muscular atrophy; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Combined treatment with the histone deacetylase inhibitor LBH589 and a splice-switch antisense oligonucleotide enhances SMN2 splicing and SMN expression in Spinal Muscular Atrophy cells.J Neurochem. 2020 Apr;153(2):264-275. doi: 10.1111/jnc.14935. Epub 2020 Jan 8. J Neurochem. 2020. PMID: 31811660
-
Improved antisense oligonucleotide design to suppress aberrant SMN2 gene transcript processing: towards a treatment for spinal muscular atrophy.PLoS One. 2013 Apr 22;8(4):e62114. doi: 10.1371/journal.pone.0062114. Print 2013. PLoS One. 2013. PMID: 23630626 Free PMC article.
-
Combination of valproic acid and morpholino splice-switching oligonucleotide produces improved outcomes in spinal muscular atrophy patient-derived fibroblasts.Neurochem Int. 2017 Sep;108:213-221. doi: 10.1016/j.neuint.2017.02.016. Epub 2017 Apr 4. Neurochem Int. 2017. PMID: 28389270
-
Splicing of the Survival Motor Neuron genes and implications for treatment of SMA.Front Biosci (Landmark Ed). 2010 Jun 1;15(3):1191-1204. doi: 10.2741/3670. Front Biosci (Landmark Ed). 2010. PMID: 20515750 Free PMC article. Review.
-
Recent Advances and Clinical Applications of Exon Inclusion for Spinal Muscular Atrophy.Methods Mol Biol. 2018;1828:57-68. doi: 10.1007/978-1-4939-8651-4_3. Methods Mol Biol. 2018. PMID: 30171534 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

