In Silico and In Vitro Screening of 50 Curcumin Compounds as EGFR and NF-κB Inhibitors
- PMID: 35409325
- PMCID: PMC9000198
- DOI: 10.3390/ijms23073966
In Silico and In Vitro Screening of 50 Curcumin Compounds as EGFR and NF-κB Inhibitors
Abstract
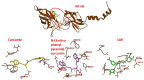
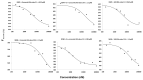
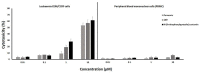
The improvement of cancer chemotherapy remains a major challenge, and thus new drugs are urgently required to develop new treatment regimes. Curcumin, a polyphenolic antioxidant derived from the rhizome of turmeric (Curcuma longa L.), has undergone extensive preclinical investigations and, thereby, displayed remarkable efficacy in vitro and in vivo against cancer and other disorders. However, pharmacological limitations of curcumin stimulated the synthesis of numerous novel curcumin analogs, which need to be evaluated for their therapeutic potential. In the present study, we calculated the binding affinities of 50 curcumin derivatives to known cancer-related target proteins of curcumin, i.e., epidermal growth factor receptor (EGFR) and nuclear factor κB (NF-κB) by using a molecular docking approach. The binding energies for EGFR were in a range of −12.12 (±0.21) to −7.34 (±0.07) kcal/mol and those for NF-κB ranged from −12.97 (±0.47) to −6.24 (±0.06) kcal/mol, indicating similar binding affinities of the curcumin compounds for both target proteins. The predicted receptor-ligand binding constants for EGFR and curcumin derivatives were in a range of 0.00013 (±0.00006) to 3.45 (±0.10) µM and for NF-κB in a range of 0.0004 (±0.0003) to 10.05 (±4.03) µM, indicating that the receptor-ligand binding was more stable for EGFR than for NF-κB. Twenty out of 50 curcumin compounds showed binding energies to NF-κB smaller than −10 kcal/mol, while curcumin as a lead compound revealed free binding energies of >−10 kcal/mol. Comparable data were obtained for EGFR: 15 out of 50 curcumin compounds were bound to EGFR with free binding energies of <−10 kcal/mol, while the binding affinity of curcumin itself was >−10 kcal/mol. This indicates that the derivatization of curcumin may indeed be a promising strategy to improve targe specificity and to obtain more effective anticancer drug candidates. The in silico results have been exemplarily validated using microscale thermophoresis. The bioactivity has been further investigated by using resazurin cell viability assay, lactate dehydrogenase assay, flow cytometric measurement of reactive oxygen species, and annexin V/propidium iodide assay. In conclusion, molecular docking represents a valuable approach to facilitate and speed up the identification of novel targeted curcumin-based drugs to treat cancer.
Keywords: bioinformatics; cancer; natural products; phytochemicals; synthetic derivatives; virtual drug screening.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures







References
-
- Toyoda Y., Tabuchi T., Nakayama T., Hojo S., Yoshioka S., Maeura Y. Past trends and future estimation of annual breast cancer incidence in Osaka, Japan. Asian Pac. J. Cancer Prev. 2016;17:52. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

