Candidate Genes and Pathways in Rice Co-Responding to Drought and Salt Identified by gcHap Network
- PMID: 35409377
- PMCID: PMC8999833
- DOI: 10.3390/ijms23074016
Candidate Genes and Pathways in Rice Co-Responding to Drought and Salt Identified by gcHap Network
Abstract
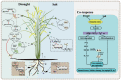
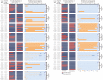
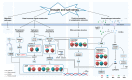
Drought and salinity stresses are significant abiotic factors that limit rice yield. Exploring the co-response mechanism to drought and salt stress will be conducive to future rice breeding. A total of 1748 drought and salt co-responsive genes were screened, most of which are enriched in plant hormone signal transduction, protein processing in the endoplasmic reticulum, and the MAPK signaling pathways. We performed gene-coding sequence haplotype (gcHap) network analysis on nine important genes out of the total amount, which showed significant differences between the Xian/indica and Geng/japonica population. These genes were combined with related pathways, resulting in an interesting mechanistic draft called the 'gcHap-network pathway'. Meanwhile, we collected a lot of drought and salt breeding varieties, especially the introgression lines (ILs) with HHZ as the parent, which contained the above-mentioned nine genes. This might imply that these ILs have the potential to improve the tolerance to drought and salt. In this paper, we focus on the relationship of drought and salt co-response gene gcHaps and their related pathways using a novel angle. The haplotype network will be helpful to explore the desired haplotypes that can be implemented in haplotype-based breeding programs.
Keywords: QTL/genes; drought tolerance; gcHap-network; pathway; rice; salt tolerance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network.Plant Sci. 2016 Jan;242:278-287. doi: 10.1016/j.plantsci.2015.08.008. Epub 2015 Aug 20. Plant Sci. 2016. PMID: 26566845
-
Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress.BMC Genomics. 2014 Nov 26;15(1):1026. doi: 10.1186/1471-2164-15-1026. BMC Genomics. 2014. PMID: 25428615 Free PMC article.
-
The landscape of gene-CDS-haplotype diversity in rice: Properties, population organization, footprints of domestication and breeding, and implications for genetic improvement.Mol Plant. 2021 May 3;14(5):787-804. doi: 10.1016/j.molp.2021.02.003. Epub 2021 Feb 10. Mol Plant. 2021. PMID: 33578043
-
Improvement of Drought Tolerance in Rice (Oryza sativa L.): Genetics, Genomic Tools, and the WRKY Gene Family.Biomed Res Int. 2018 Aug 7;2018:3158474. doi: 10.1155/2018/3158474. eCollection 2018. Biomed Res Int. 2018. PMID: 30175125 Free PMC article. Review.
-
Stacking for future: Pyramiding genes to improve drought and salinity tolerance in rice.Physiol Plant. 2021 Jun;172(2):1352-1362. doi: 10.1111/ppl.13270. Epub 2020 Nov 21. Physiol Plant. 2021. PMID: 33180968 Review.
Cited by
-
Physiological and molecular implications of multiple abiotic stresses on yield and quality of rice.Front Plant Sci. 2023 Jan 11;13:996514. doi: 10.3389/fpls.2022.996514. eCollection 2022. Front Plant Sci. 2023. PMID: 36714754 Free PMC article. Review.
-
Genomics and transcriptomics to protect rice (Oryza sativa. L.) from abiotic stressors: -pathways to achieving zero hunger.Front Plant Sci. 2022 Oct 20;13:1002596. doi: 10.3389/fpls.2022.1002596. eCollection 2022. Front Plant Sci. 2022. PMID: 36340401 Free PMC article. Review.
-
Combined transcriptome and metabolome reveal glutathione metabolism plays a critical role in resistance to salinity in rice landraces HD961.Front Plant Sci. 2022 Sep 7;13:952595. doi: 10.3389/fpls.2022.952595. eCollection 2022. Front Plant Sci. 2022. PMID: 36160959 Free PMC article.
References
-
- Jing R.L. Advances of research on drought resistance and water use efficiency in crop plants. Rev. China Agric. Sci. Technol. 2007;1:1–5.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

