shRNAs Targeting a Common KCNQ1 Variant Could Alleviate Long-QT1 Disease Severity by Inhibiting a Mutant Allele
- PMID: 35409410
- PMCID: PMC9000197
- DOI: 10.3390/ijms23074053
shRNAs Targeting a Common KCNQ1 Variant Could Alleviate Long-QT1 Disease Severity by Inhibiting a Mutant Allele
Abstract
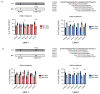
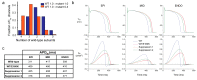
Long-QT syndrome type 1 (LQT1) is caused by mutations in KCNQ1. Patients heterozygous for such a mutation co-assemble both mutant and wild-type KCNQ1-encoded subunits into tetrameric Kv7.1 potassium channels. Here, we investigated whether allele-specific inhibition of mutant KCNQ1 by targeting a common variant can shift the balance towards increased incorporation of the wild-type allele to alleviate the disease in human-induced pluripotent stem-cell-derived cardiomyocytes (hiPSC-CMs). We identified the single nucleotide polymorphisms (SNP) rs1057128 (G/A) in KCNQ1, with a heterozygosity of 27% in the European population. Next, we determined allele-specificity of short-hairpin RNAs (shRNAs) targeting either allele of this SNP in hiPSC-CMs that carry an LQT1 mutation. Our shRNAs downregulated 60% of the A allele and 40% of the G allele without affecting the non-targeted allele. Suppression of the mutant KCNQ1 allele by 60% decreased the occurrence of arrhythmic events in hiPSC-CMs measured by a voltage-sensitive reporter, while suppression of the wild-type allele increased the occurrence of arrhythmic events. Furthermore, computer simulations based on another LQT1 mutation revealed that 60% suppression of the mutant KCNQ1 allele shortens the prolonged action potential in an adult cardiomyocyte model. We conclude that allele-specific inhibition of a mutant KCNQ1 allele by targeting a common variant may alleviate the disease. This novel approach avoids the need to design shRNAs to target every single mutation and opens up the exciting possibility of treating multiple LQT1-causing mutations with only two shRNAs.
Keywords: RNA interference; arrhythmia; gene therapy; hiPSC-cardiomyocytes; long-QT syndrome type 1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Goldenberg I., Horr S., Moss A.J., Lopes C.M., Barsheshet A., McNitt S., Zareba W., Andrews M.L., Robinson J.L., Locati E.H., et al. Risk for life-threatening cardiac events in patients with genotype-confirmed long-QT syndrome and normal-range corrected QT intervals. J. Am. Coll. Cardiol. 2011;57:51–59. doi: 10.1016/j.jacc.2010.07.038. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

