The local burden of disease during the first wave of the COVID-19 epidemic in England: estimation using different data sources from changing surveillance practices
- PMID: 35410184
- PMCID: PMC8996221
- DOI: 10.1186/s12889-022-13069-0
The local burden of disease during the first wave of the COVID-19 epidemic in England: estimation using different data sources from changing surveillance practices
Erratum in
-
Correction: The local burden of disease during the first wave of the COVID-19 epidemic in England: estimation using different data sources from changing surveillance practices.BMC Public Health. 2022 Jun 7;22(1):1140. doi: 10.1186/s12889-022-13320-8. BMC Public Health. 2022. PMID: 35672722 Free PMC article. No abstract available.
Abstract
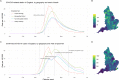
Background: The COVID-19 epidemic has differentially impacted communities across England, with regional variation in rates of confirmed cases, hospitalisations and deaths. Measurement of this burden changed substantially over the first months, as surveillance was expanded to accommodate the escalating epidemic. Laboratory confirmation was initially restricted to clinical need ("pillar 1") before expanding to community-wide symptomatics ("pillar 2"). This study aimed to ascertain whether inconsistent measurement of case data resulting from varying testing coverage could be reconciled by drawing inference from COVID-19-related deaths.
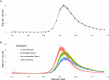
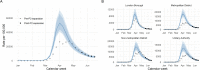
Methods: We fit a Bayesian spatio-temporal model to weekly COVID-19-related deaths per local authority (LTLA) throughout the first wave (1 January 2020-30 June 2020), adjusting for the local epidemic timing and the age, deprivation and ethnic composition of its population. We combined predictions from this model with case data under community-wide, symptomatic testing and infection prevalence estimates from the ONS infection survey, to infer the likely trajectory of infections implied by the deaths in each LTLA.
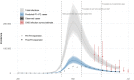
Results: A model including temporally- and spatially-correlated random effects was found to best accommodate the observed variation in COVID-19-related deaths, after accounting for local population characteristics. Predicted case counts under community-wide symptomatic testing suggest a total of 275,000-420,000 cases over the first wave - a median of over 100,000 additional to the total confirmed in practice under varying testing coverage. This translates to a peak incidence of around 200,000 total infections per week across England. The extent to which estimated total infections are reflected in confirmed case counts was found to vary substantially across LTLAs, ranging from 7% in Leicester to 96% in Gloucester with a median of 23%.
Conclusions: Limitations in testing capacity biased the observed trajectory of COVID-19 infections throughout the first wave. Basing inference on COVID-19-related mortality and higher-coverage testing later in the time period, we could explore the extent of this bias more explicitly. Evidence points towards substantial under-representation of initial growth and peak magnitude of infections nationally, to which different parts of the country contribute unequally.
© 2022. The Author(s).
Conflict of interest statement
The author(s) declare(s) that they have no competing interests.
Figures


References
-
- Holden B, Quinney A, Padfield S, Morton W, Coles S, Manley P, et al. COVID-19: public health management of the first two confirmed cases identified in the UK. Epidemiol Infect. 2020;148 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7484301/. - PMC - PubMed
-
- Coronavirus (COVID-19): scaling up testing programmes [Internet]. GOV.UK. Available from: https://www.gov.uk/government/publications/coronavirus-covid-19-scaling-.... Accessed 14 May 2021.
-
- Deaths involving COVID-19 by local area and socioeconomic deprivation - Office for National Statistics [Internet]. Available from: https://www.ons.gov.uk/peoplepopulationandcommunity/birthsdeathsandmarri.... Accessed 17 Mar 2021.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

