Single-cell analysis of somatic mutations in human bronchial epithelial cells in relation to aging and smoking
- PMID: 35410377
- PMCID: PMC9844147
- DOI: 10.1038/s41588-022-01035-w
Single-cell analysis of somatic mutations in human bronchial epithelial cells in relation to aging and smoking
Abstract
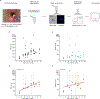
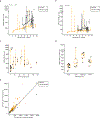
Although lung cancer risk among smokers is dependent on smoking dose, it remains unknown if this increased risk reflects an increased rate of somatic mutation accumulation in normal lung cells. Here, we applied single-cell whole-genome sequencing of proximal bronchial basal cells from 33 participants aged between 11 and 86 years with smoking histories varying from never-smoking to 116 pack-years. We found an increase in the frequency of single-nucleotide variants and small insertions and deletions with chronological age in never-smokers, with mutation frequencies significantly elevated among smokers. When plotted against smoking pack-years, mutations followed the linear increase in cancer risk until about 23 pack-years, after which no further increase in mutation frequency was observed, pointing toward individual selection for mutation avoidance. Known lung cancer-defined mutation signatures tracked with both age and smoking. No significant enrichment for somatic mutations in lung cancer driver genes was observed.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
A.Y.M., X.D., and J.V. are cofounders of SingulOmics Corp. The remaining authors declare no other competing interests.
Figures






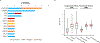

References
-
- Flanders WD, Lally CA, Zhu B-P, Henley SJ & Thun MJ Lung Cancer Mortality in Relation to Age, Duration of Smoking, and Daily Cigarette Consumption. Cancer Research 63(2003). - PubMed
-
- Spivack SD, Fasco MJ, Walker VE & Kaminsky LS The molecular epidemiology of lung cancer. Crit Rev Toxicol 27, 319–65 (1997). - PubMed
Methods-only References:
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

