Transcriptome analysis of adipocytokines and their-related LncRNAs in lung adenocarcinoma revealing the association with prognosis, immune infiltration, and metabolic characteristics
- PMID: 35410586
- PMCID: PMC9037474
- DOI: 10.1080/21623945.2022.2064956
Transcriptome analysis of adipocytokines and their-related LncRNAs in lung adenocarcinoma revealing the association with prognosis, immune infiltration, and metabolic characteristics
Abstract
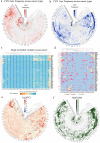
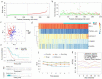
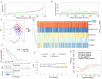
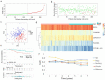
Lung adenocarcinoma (LUAD) is amongst the major contributors to cancer-related deaths on a global scale. Adipocytokines and long non-coding RNAs (lncRNAs) are indispensable participants in cancer. We performed a pan-cancer analysis of the mRNA expression, single nucleotide variation, copy number variation, and prognostic value of adipocytokines. LUAD samples were obtained from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases. Simultaneously, train, internal and external cohorts were grouped. After a stepwise screening of optimized genes through least absolute shrinkage and selection operator regression analysis, random forest algorithm,, and Cox regression analysis, an adipocytokine-related prognostic signature (ARPS) with superior performance compared with four additional well-established signatures for survival prediction was constructed. After determination of risk levels, the discrepancy of immune microenvironment, immune checkpoint gene expression, immune subtypes, and immune response in low- and high-risk cohorts were explored through multiple bioinformatics methods. Abnormal pathways underlying high- and low-risk subgroups were identified through gene set enrichment analysis (GSEA). Immune-and metabolism-related pathways that were correlated with risk score were selected through single sample GSEA. Finally, a nomogram with satisfied predictive survival probability was plotted. In summary, this study offers meaningful information for clinical treatment and scientific investigation.
Keywords: Lung adenocarcinoma; adipocytokine; immune infiltration; long non-coding RNA; prognostic signature.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





References
-
- Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
