Stratification of radiosensitive brain metastases based on an actionable S100A9/RAGE resistance mechanism
- PMID: 35411077
- PMCID: PMC9018424
- DOI: 10.1038/s41591-022-01749-8
Stratification of radiosensitive brain metastases based on an actionable S100A9/RAGE resistance mechanism
Abstract
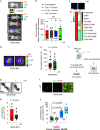
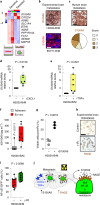
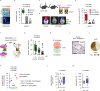
Whole-brain radiotherapy (WBRT) is the treatment backbone for many patients with brain metastasis; however, its efficacy in preventing disease progression and the associated toxicity have questioned the clinical impact of this approach and emphasized the need for alternative treatments. Given the limited therapeutic options available for these patients and the poor understanding of the molecular mechanisms underlying the resistance of metastatic lesions to WBRT, we sought to uncover actionable targets and biomarkers that could help to refine patient selection. Through an unbiased analysis of experimental in vivo models of brain metastasis resistant to WBRT, we identified activation of the S100A9-RAGE-NF-κB-JunB pathway in brain metastases as a potential mediator of resistance in this organ. Targeting this pathway genetically or pharmacologically was sufficient to revert the WBRT resistance and increase therapeutic benefits in vivo at lower doses of radiation. In patients with primary melanoma, lung or breast adenocarcinoma developing brain metastasis, endogenous S100A9 levels in brain lesions correlated with clinical response to WBRT and underscored the potential of S100A9 levels in the blood as a noninvasive biomarker. Collectively, we provide a molecular framework to personalize WBRT and improve its efficacy through combination with a radiosensitizer that balances therapeutic benefit and toxicity.
© 2022. The Author(s).
Conflict of interest statement
R.L. receives research funding from Bristol Myers Squibb, Pierre Fabre and AstraZeneca; P.L. serves as a paid advisor/speaker for Bristol Myers Squibb, Merck Sharp and Dohme, Roche, Novartis, Amgen, Pierre Fabre, Nektar and MelaGenix. P.L. receives travel support from Bristol Myers Squibb and Merck Sharp and Dohme. P.L. receives research support from Bristol Myers Squibb and Pierre Fabre. The other authors declare no competing interests.
Figures






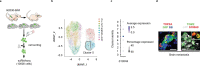
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

