Genomic Insights Into the Mechanism of Carbapenem Resistance Dissemination in Enterobacterales From a Tertiary Public Heath Setting in South Asia
- PMID: 35412593
- PMCID: PMC9825829
- DOI: 10.1093/cid/ciac287
Genomic Insights Into the Mechanism of Carbapenem Resistance Dissemination in Enterobacterales From a Tertiary Public Heath Setting in South Asia
Abstract
Summary: 10.6% patients were CRE positive. Only 27% patients were prescribed at least 1 antibiotic to which infecting pathogen was susceptible. Burn and ICU admission and antibiotics exposures facilitate CRE acquisition. Escherichia coli ST167 was the dominant CRE clone.
Background: Given the high prevalence of multidrug resistance (MDR) across South Asian (SA) hospitals, we documented the epidemiology of carbapenem-resistant Enterobacterales (CRE) infections at Dhaka Medical College Hospital between October 2016 and September 2017.
Methods: We enrolled patients and collected epidemiology and outcome data. All Enterobacterales were characterized phenotypically and by whole-genome sequencing. Risk assessment for the patients with CRE was performed compared with patients with carbapenem-susceptible Enterobacterales (CSE).
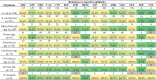
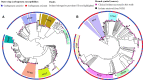
Results: 10.6% of all 1831 patients with a clinical specimen collected had CRE. In-hospital 30-day mortality was significantly higher with CRE [50/180 (27.8%)] than CSE [42/312 (13.5%)] (P = .001); however, for bloodstream infections, this was nonsignificant. Of 643 Enterobacterales isolated, 210 were CRE; blaNDM was present in 180 isolates, blaOXA-232 in 26, blaOXA-181 in 24, and blaKPC-2 in 5. Despite this, ceftriaxone was the most commonly prescribed empirical antibiotic and only 27% of patients were prescribed at least 1 antibiotic to which their infecting pathogen was susceptible. Significant risk factors for CRE isolation included burns unit and intensive care unit admission, and prior exposure to levofloxacin, amikacin, clindamycin, and meropenem. Escherichia coli ST167 was the dominant CRE clone. Clustering suggested clonal transmission of Klebsiella pneumoniae ST15 and the MDR hypervirulent clone, ST23. The major trajectories involved in horizontal gene transfer were IncFII and IncX3, IS26, and Tn3.
Conclusions: This is the largest study from an SA public hospital combining outcome, microbiology, and genomics. The findings indicate the urgent implementation of targeted diagnostics, appropriate antibiotic use, and infection-control interventions in SA public institutions.
Keywords: Bangladesh; South Asia; carbapenem-resistant Enterobacterales; outbreak; plasmid-mediated resistance.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
Figures






References
-
- World Health Organization . WHO publishes list of bacteria for which new antibiotics are urgently needed. Available at:https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-ba.... Accessed 10 November 2020.
-
- Mirande C, Bizine I, Giannetti A, Picot N, van Belkum A. Epidemiological aspects of healthcare-associated infections and microbial genomics. Eur J Clin Microbiol Infect Dis 2018; 37:823–31. - PubMed
-
- Stewardson AJ, Marimuthu K, Sengupta S, et al. . Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study. Lancet Infect Dis 2019; 19:601–10. - PubMed

