Visualization of ligand-induced c-MYC duplex-quadruplex transition and direct exploration of the altered c-MYC DNA-protein interactions in cells
- PMID: 35412611
- PMCID: PMC9071431
- DOI: 10.1093/nar/gkac245
Visualization of ligand-induced c-MYC duplex-quadruplex transition and direct exploration of the altered c-MYC DNA-protein interactions in cells
Abstract
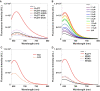
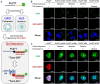
Ligand-Induced duplex-quadruplex transition within the c-MYC promoter region is one of the most studied and advanced ideas for c-MYC regulation. Despite its importance, there is a lack of methods for monitoring such process in cells, hindering a better understanding of the essence of c-MYC G-quadruplex as a drug target. Here we developed a new fluorescent probe ISCH-MYC for specific c-MYC G-quadruplex recognition based on GTFH (G-quadruplex-Triggered Fluorogenic Hybridization) strategy. We validated that ISCH-MYC displayed distinct fluorescence enhancement upon binding to c-MYC G-quadruplex, which allowed the duplex-quadruplex transition detection of c-MYC G-rich DNA in cells. Using ISCH-MYC, we successfully characterized the induction of duplex to G-quadruplex transition in the presence of G-quadruplex stabilizing ligand PDS and further monitored and evaluated the altered interactions of relevant transcription factors Sp1 and CNBP with c-MYC G-rich DNA. Thus, our study provides a visualization strategy to explore the mechanism of G-quadruplex stabilizing ligand action on c-MYC G-rich DNA and relevant proteins, thereby empowering future drug discovery efforts targeting G-quadruplexes.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

