WashU Epigenome Browser update 2022
- PMID: 35412637
- PMCID: PMC9252771
- DOI: 10.1093/nar/gkac238
WashU Epigenome Browser update 2022
Abstract
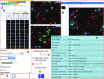
WashU Epigenome Browser (https://epigenomegateway.wustl.edu/browser/) is a web-based genomic data exploration tool that provides visualization, integration, and analysis of epigenomic datasets. The newly renovated user interface and functions have enabled researchers to engage with the browser and genomic data more efficiently and effectively since 2018. Here, we introduce a new integrated panel design in the browser that allows users to interact with 1D (genomic features), 2D (such as Hi-C), 3D (genome structure), and 4D (time series) data in a single web page. The browser can display three-dimensional chromatin structures with the 3D viewer module. The 4D tracks, called 'Dynamic' tracks, animatedly display time-series data, allowing for a more striking visual impact to identify the gene or genomic region candidates as a function of time. Genomic data, such as annotation features, numerical values, and chromatin interaction data can all be viewed in the dynamic track mode. Imaging data from microscopy experiments can also be displayed in the browser. In addition to software development, we continue to service and expand the data hubs we host for large consortia including 4DN, Roadmap Epigenomics, TaRGET and ENCODE, among others. Our growing user/developer community developed additional track types as plugins, such as qBed and dynseq tracks, which extend the utility of the browser. The browser serves as a foundation for additional genomics platforms including the WashU Virus Genome Browser (for COVID-19 research) and the Comparative Genome Browser. The WashU Epigenome Browser can also be accessed freely through Amazon Web Services at https://epigenomegateway.org/.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Encode Project Consortium The ENCODE (ENCyclopedia of DNA elements) project. Science. 2004; 306:636–640. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

