Elucidation of CKAP4-remodeled cell mechanics in driving metastasis of bladder cancer through aptamer-based target discovery
- PMID: 35412892
- PMCID: PMC9169774
- DOI: 10.1073/pnas.2110500119
Elucidation of CKAP4-remodeled cell mechanics in driving metastasis of bladder cancer through aptamer-based target discovery
Abstract
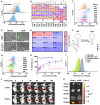
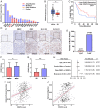
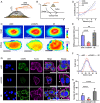
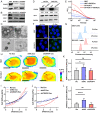
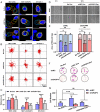
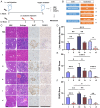
Metastasis contributes to the dismal prognosis of bladder cancer (BLCA). The mechanical status of the cell membrane is expected to mirror the ability of cell migration to promote cancer metastasis. However, the mechanical characteristics and underlying molecular profile associated with BLCA metastasis remain obscure. To study the unique cellular architecture and traits associated with cell migration, using a process called cell-based systematic evolution of ligands by exponential enrichment (cell-SELEX) we generated an aptamer-based molecular probe, termed spl3c, which identified cytoskeleton-associated protein 4 (CKAP4). CKAP4 was associated with tumor metastasis in BLCA, but we also found it to be a mechanical regulator of BLCA cells through the maintenance of a central-to-peripheral gradient of stiffness on the cell membrane. Notably, such mechanical traits were transportable through exosome-mediated intercellular CKAP4 trafficking, leading to significant enhancement of migration in recipient cells and, consequently, aggravating metastatic potential in vivo. Taken together, our study shows the robustness of this aptamer-based molecular tool for biomarker discovery, revealing the dominance of a CKAP4-induced central-to-peripheral gradient of membrane stiffness that benefits cell migration and delineating the role of exosomes in mediating mechanical signaling in BLCA metastasis.
Keywords: CKAP4; aptamer; bladder cancer; cell mechanics; cell migration.
Conflict of interest statement
The authors declare no competing interest.
Figures

Comment in
-
Uro-Science.J Urol. 2022 Dec;208(6):1340-1342. doi: 10.1097/JU.0000000000002980. Epub 2022 Sep 26. J Urol. 2022. PMID: 36154669 No abstract available.
References
-
- Massou S., et al. , Cell stretching is amplified by active actin remodelling to deform and recruit proteins in mechanosensitive structures. Nat. Cell Biol. 22, 1011–1023 (2020). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

