Imprinted SARS-CoV-2-specific memory lymphocytes define hybrid immunity
- PMID: 35413241
- PMCID: PMC8926873
- DOI: 10.1016/j.cell.2022.03.018
Imprinted SARS-CoV-2-specific memory lymphocytes define hybrid immunity
Abstract
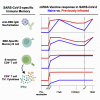
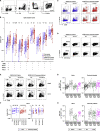
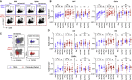
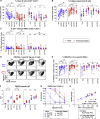
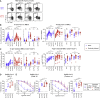
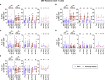
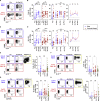
Immune memory is tailored by cues that lymphocytes perceive during priming. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic created a situation in which nascent memory could be tracked through additional antigen exposures. Both SARS-CoV-2 infection and vaccination induce multifaceted, functional immune memory, but together, they engender improved protection from disease, termed hybrid immunity. We therefore investigated how vaccine-induced memory is shaped by previous infection. We found that following vaccination, previously infected individuals generated more SARS-CoV-2 RBD-specific memory B cells and variant-neutralizing antibodies and a distinct population of IFN-γ and IL-10-expressing memory SARS-CoV-2 spike-specific CD4+ T cells than previously naive individuals. Although additional vaccination could increase humoral memory in previously naive individuals, it did not recapitulate the distinct CD4+ T cell cytokine profile observed in previously infected subjects. Thus, imprinted features of SARS-CoV-2-specific memory lymphocytes define hybrid immunity.
Keywords: COVID-19; SARS-CoV-2; adaptive immune response; human; hybrid Immunity; memory B cell; memory T cell; vaccine.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.P. is a member of the Scientific Advisory Board of VaxArt and NeoLeukin Inc.
Figures





References
-
- Abu-Raddad L.J., Chemaitelly H., Ayoub H.H., Yassine H.M., Benslimane F.M., Khatib H.A.A., Tang P., Hasan M.R., Coyle P., Kanaani Z.A., et al. Protection afforded by the BNT162b2 and mRNA-1273 COVID-19 vaccines in fully vaccinated cohorts with and without prior infection. Preprint at medRxiv. 2021 2021.07.25.21261093.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

