Structural basis for llama nanobody recognition and neutralization of HIV-1 at the CD4-binding site
- PMID: 35413243
- PMCID: PMC9177634
- DOI: 10.1016/j.str.2022.03.012
Structural basis for llama nanobody recognition and neutralization of HIV-1 at the CD4-binding site
Abstract
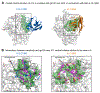
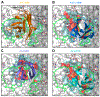
Nanobodies can achieve remarkable neutralization of genetically diverse pathogens, including HIV-1. To gain insight into their recognition, we determined crystal structures of four llama nanobodies (J3, A12, C8, and D7), all of which targeted the CD4-binding site, in complex with the HIV-1 envelope (Env) gp120 core, and determined a cryoelectron microscopy (cryo-EM) structure of J3 with the Env trimer. Crystal and cryo-EM structures of J3 complexes revealed this nanobody to mimic binding to the prefusion-closed trimer for the primary site of CD4 recognition as well as a secondary quaternary site. In contrast, crystal structures of A12, C8, and D7 with gp120 revealed epitopes that included portions of the gp120 inner domain, inaccessible on the prefusion-closed trimer. Overall, these structures explain the broad and potent neutralization of J3 and limited neutralization of A12, C8, and D7, which utilized binding modes incompatible with the neutralization-targeted prefusion-closed conformation of Env.
Keywords: CD4-binding site; HIV; cryo-EM; crystal structure; envelope trimer; llama VHH; nanobody; neutralization; single-domain antibody; steric clash.
Published by Elsevier Ltd.
Conflict of interest statement
Declaration of interests The authors declare no competing interest.
Figures




References
-
- Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, et al. (2004). Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat 11, 53–55. - PubMed
-
- Agirre J, Iglesias-Fernandez J, Rovira C, Davies GJ, Wilson KS, and Cowtan KD (2015). Privateer: software for the conformational validation of carbohydrate structures. Nat Struct Mol Biol 22, 833–834. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

