Growth factor independence underpins a paroxysmal, aggressive Wnt5aHigh/EphA2Low phenotype in glioblastoma stem cells, conducive to experimental combinatorial therapy
- PMID: 35414102
- PMCID: PMC9004109
- DOI: 10.1186/s13046-022-02333-1
Growth factor independence underpins a paroxysmal, aggressive Wnt5aHigh/EphA2Low phenotype in glioblastoma stem cells, conducive to experimental combinatorial therapy
Abstract
Background: Glioblastoma multiforme (GBM) is an incurable tumor, with a median survival rate of only 14-15 months. Along with heterogeneity and unregulated growth, a central matter in dealing with GBMs is cell invasiveness. Thus, improving prognosis requires finding new agents to inhibit key multiple pathways, even simultaneously. A subset of GBM stem-like cells (GSCs) may account for tumorigenicity, representing, through their pathways, the proper cellular target in the therapeutics of glioblastomas. GSCs cells are routinely enriched and expanded due to continuous exposure to specific growth factors, which might alter some of their intrinsic characteristic and hide therapeutically relevant traits.
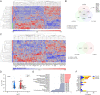
Methods: By removing exogenous growth factors stimulation, here we isolated and characterized a subset of GSCs with a "mitogen-independent" phenotype (I-GSCs) from patient's tumor specimens. Differential side-by-side comparative functional and molecular analyses were performed either in vitro or in vivo on these cells versus their classical growth factor (GF)-dependent counterpart (D-GSCs) as well as their tissue of origin. This was performed to pinpoint the inherent GSCs' critical regulators, with particular emphasis on those involved in spreading and tumorigenic potential. Transcriptomic fingerprints were pointed out by ANOVA with Benjamini-Hochberg False Discovery Rate (FDR) and association of copy number alterations or somatic mutations was determined by comparing each subgroup with a two-tailed Fisher's exact test. The combined effects of interacting in vitro and in vivo with two emerging GSCs' key regulators, such as Wnt5a and EphA2, were then predicted under in vivo experimental settings that are conducive to clinical applications. In vivo comparisons were carried out in mouse-human xenografts GBM model by a hierarchical linear model for repeated measurements and Dunnett's multiple comparison test with the distribution of survival compared by Kaplan-Meier method.
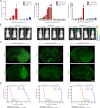
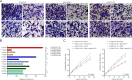
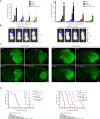
Results: Here, we assessed that a subset of GSCs from high-grade gliomas is self-sufficient in the activation of regulatory growth signaling. Furthermore, while constitutively present within the same GBM tissue, these GF-independent GSCs cells were endowed with a distinctive functional and molecular repertoire, defined by highly aggressive Wnt5aHigh/EphA2Low profile, as opposed to Wnt5aLow/EphA2High expression in sibling D-GSCs. Regardless of their GBM subtype of origin, I-GSCs, are endowed with a raised in vivo tumorigenic potential than matched D-GSCs, which were fast-growing ex-vivo but less lethal and invasive in vivo. Also, the malignant I-GSCs' transcriptomic fingerprint faithfully mirrored the original tumor, bringing into evidence key regulators of invasiveness, angiogenesis and immuno-modulators, which became candidates for glioma diagnostic/prognostic markers and therapeutic targets. Particularly, simultaneously counteracting the activity of the tissue invasive mediator Wnt5a and EphA2 tyrosine kinase receptor addictively hindered GSCs' tumorigenic and invasive ability, thus increasing survival.
Conclusion: We show how the preservation of a mitogen-independent phenotype in GSCs plays a central role in determining the exacerbated tumorigenic and high mobility features distinctive of GBM. The exploitation of the I-GSCs' peculiar features shown here offers new ways to identify novel, GSCs-specific effectors, whose modulation can be used in order to identify novel, potential molecular therapeutic targets. Furthermore, we show how the combined use of PepA, the anti-Wnt5a drug, and of ephrinA1-Fc to can hinder GSCs' lethality in a clinically relevant xenogeneic in vivo model thus being conducive to perspective, novel combinatorial clinical application.
Keywords: Anti-GSCs patient-tailored strategies; GBM cancer stem cells (GSCs); GSCs biology and biomarkers; Glioblastoma; Mitogen-independence.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests. AL Vescovi has ownership interest in Hyperstem SA.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

