Genome-wide and transcriptome-wide association studies of mammographic density phenotypes reveal novel loci
- PMID: 35414113
- PMCID: PMC9006574
- DOI: 10.1186/s13058-022-01524-0
Genome-wide and transcriptome-wide association studies of mammographic density phenotypes reveal novel loci
Abstract
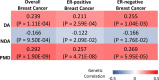
Background: Mammographic density (MD) phenotypes, including percent density (PMD), area of dense tissue (DA), and area of non-dense tissue (NDA), are associated with breast cancer risk. Twin studies suggest that MD phenotypes are highly heritable. However, only a small proportion of their variance is explained by identified genetic variants.
Methods: We conducted a genome-wide association study, as well as a transcriptome-wide association study (TWAS), of age- and BMI-adjusted DA, NDA, and PMD in up to 27,900 European-ancestry women from the MODE/BCAC consortia.
Results: We identified 28 genome-wide significant loci for MD phenotypes, including nine novel signals (5q11.2, 5q14.1, 5q31.1, 5q33.3, 5q35.1, 7p11.2, 8q24.13, 12p11.2, 16q12.2). Further, 45% of all known breast cancer SNPs were associated with at least one MD phenotype at p < 0.05. TWAS further identified two novel genes (SHOX2 and CRISPLD2) whose genetically predicted expression was significantly associated with MD phenotypes.
Conclusions: Our findings provided novel insight into the genetic background of MD phenotypes, and further demonstrated their shared genetic basis with breast cancer.
Keywords: Breast cancer; Genome-wide association study (GWAS); Mammographic density; Transcriptome-wide association study (TWAS).
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

