LC3 subfamily in cardiolipin-mediated mitophagy: a comparison of the LC3A, LC3B and LC3C homologs
- PMID: 35414338
- PMCID: PMC9673933
- DOI: 10.1080/15548627.2022.2062111
LC3 subfamily in cardiolipin-mediated mitophagy: a comparison of the LC3A, LC3B and LC3C homologs
Abstract
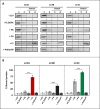
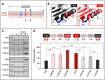
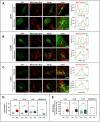
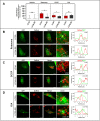
Externalization of the phospholipid cardiolipin (CL) to the outer mitochondrial membrane has been proposed to act as a mitophagy trigger. CL would act as a signal for binding the LC3 macroautophagy/autophagy proteins. As yet, the behavior of the LC3-subfamily members has not been directly compared in a detailed way. In the present contribution, an analysis of LC3A, LC3B and LC3C interaction with CL-containing model membranes, and of their ability to translocate to mitochondria, is described. Binding of LC3A to CL was stronger than that of LC3B; both proteins showed a similar ability to colocalize with mitochondria upon induction of CL externalization in SH-SY5Y cells. Besides, the double silencing of LC3A and LC3B proteins was seen to decrease CCCP-induced mitophagy. Residues 14 and 18 located in the N-terminal region of LC3A were shown to be important for its recognition of damaged mitochondria during rotenone- or CCCP-induced mitophagy. Moreover, the in vitro results suggested a possible role of LC3A, but not of LC3B, in oxidized-CL recognition as a counterweight to excessive apoptosis activation. In the case of LC3C, even if this protein showed a stronger CL binding than LC3B or LC3A, the interaction was less specific, and colocalization of LC3C with mitochondria was not rotenone dependent. These results suggest that, at variance with LC3A, LC3C does not participate in cargo recognition during CL-mediated-mitophagy. The data support the notion that the various LC3-subfamily members might play different roles during autophagy initiation, identifying LC3A as a novel stakeholder in CL-mediated mitophagy. Abbreviations: ACTB/β-actin: actin beta; Atg8: autophagy-related 8; CL: cardiolipin; CCCP: carbonyl cyanide m-chlorophenyl hydrazone; DMSO: dimethyl sulfoxide; DOPE: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; DTT: DL-dithiothreitol; FKBP8: FKBP prolyl isomerase 8; GABARAP: GABA type A receptor associated protein; GABARAPL1: GABA type A receptor associated protein like 1; GABARAPL2: GABA type A receptor associated protein like 2; GFP: green fluorescent protein; IMM: inner mitochondrial membrane; LUV/LUVs: large unilamellar vesicle/s; MAP1LC3A/LC3A: microtubule associated protein 1 light chain 3 alpha; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; MAP1LC3C/LC3C: microtubule associated protein 1 light chain 3 gamma; NME4/NDPK-D/Nm23-H4: NME/NM23 nucleoside diphosphate kinase 4; O/A: oligomycin A + antimycin A; OMM: outer mitochondrial membrane; PA: phosphatidic acid; PC: phosphatidylcholine; PG: phosphatidylglycerol; PINK1: PTEN induced putative kinase 1; PtdIns4P: phosphatidylinositol-4-phosphate; Rho-PE: lissamine rhodamine phosphatidylethanolamine; SUV/SUVs: small unilamellar vesicle/s.
Keywords: Atg8; LC3/GABARAP-protein family; autophagosome; autophagy cargo recognition; lipid oxidation; lipid-protein interaction; membrane curvature; mitochondria; negatively charged phospholipids.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





References
-
- Schaaf MBE, Keulers TG, Vooijs MA, et al. LC3/GABARAP family proteins: autophagy‐(un)related functions. FASEB J. 2016;30:3961–3978. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
