Plasmid Viability Depends on the Ecological Setting of Hosts within a Multiplasmid Community
- PMID: 35416702
- PMCID: PMC9045312
- DOI: 10.1128/spectrum.00133-22
Plasmid Viability Depends on the Ecological Setting of Hosts within a Multiplasmid Community
Abstract
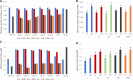
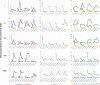
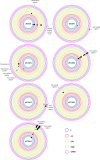
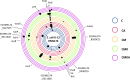
Plasmids are extrachromosomal genetic elements, some of which disperse horizontally between different strains and species of bacteria. They are a major factor in the dissemination of virulence factors and antibiotic resistance. Understanding the ecology of plasmids has a notable anthropocentric value, and therefore, the interactions between bacterial hosts and individual plasmids have been studied in detail. However, bacterial systems often carry multiple genetically distinct plasmids, but dynamics within these multiplasmid communities have remained unstudied. Here, we set to investigate the survival of 11 mobilizable or conjugative plasmids under five different conditions where the hosts had a differing ecological status in comparison to other bacteria in the system. The key incentive was to determine whether plasmid dynamics are reproducible and whether there are tradeoffs in plasmid fitness that stem from the ecological situation of their initial hosts. Growth rates and maximum population densities increased in all communities and treatments over the 42-day evolution experiment, although plasmid contents at the end varied notably. Large multiresistance-conferring plasmids were unfit when the community also contained smaller plasmids with fewer resistance genes. This suggests that restraining the use of a few antibiotics can make bacterial communities sensitive to others. In general, the presence or absence of antibiotic selection and plasmid-free hosts (of various fitnesses) has a notable influence on which plasmids survive. These tradeoffs in different settings can help explain, for example, why some resistance plasmids have an advantage during a rapid proliferation of antibiotic-sensitive pathogens whereas others dominate in alternative situations. IMPORTANCE Conjugative and mobilizable plasmids are ubiquitous in bacterial systems. Several different plasmids can compete within a single bacterial community. We here show that the ecological setting of the host bacteria has a notable effect on the survival of individual plasmids. Selection for opportunistic genes such as antibiotic resistance genes and the presence of plasmid-free hosts can determine which plasmids survive in the system. Host bacteria appear to adapt specifically to a situation where there are multiple plasmids present instead of alleviating the plasmid-associated fitness costs of individual plasmids. Plasmids providing antibiotic resistance survived under all conditions even if there was a constant migration of higher-fitness plasmid-free hosts and no selection via antibiotics. This study is one of the first to observe the behavior of multiple genetically different plasmids as a part of a single system.
Keywords: antibiotic resistance; multiresistance; plasmid ecology; plasmid evolution; plasmid stability; plasmid-mediated resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Decano AG, Tran N, Al-Foori H, Al-Awadi B, Campbell L, Ellison K, Mirabueno LP, Nelson M, Power S, Smith G, Smyth C, Vance Z, Woods C, Rahm A, Downing T. 2020. Plasmids shape the diverse accessory resistomes of Escherichia coli ST131. Access Microbiol 3:acmi000179. doi:10.1099/acmi.0.000179. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

