Aurintricarboxylic acid is a canonical disruptor of the TAZ-TEAD transcriptional complex
- PMID: 35417479
- PMCID: PMC9007350
- DOI: 10.1371/journal.pone.0266143
Aurintricarboxylic acid is a canonical disruptor of the TAZ-TEAD transcriptional complex
Abstract
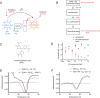
Disrupting the formation of the oncogenic YAP/TAZ-TEAD transcriptional complex holds substantial therapeutic potential. However, the three protein interaction interfaces of this complex cannot be easily disrupted using small molecules. Here, we report that the pharmacologically active small molecule aurintricarboxylic acid (ATA) acts as a disruptor of the TAZ-TEAD complex. ATA was identified in a high-throughput screen using a TAZ-TEAD AlphaLISA assay that was tailored to identify disruptors of this transcriptional complex. We further used fluorescence polarization assays both to confirm disruption of the TAZ-TEAD complex and to demonstrate that ATA binds to interface 3. We have previously shown that cell-based models that express the oncogenic TAZ-CAMTA1 (TC) fusion protein display enhanced TEAD transcriptional activity because TC functions as an activated form of TAZ. Utilizing cell-based studies and our TC model system, we performed TC/TEAD reporter, RNA-Seq, and qPCR assays and found that ATA inhibits TC/TEAD transcriptional activity. Further, disruption of TC/TEAD and TAZ/TEAD interaction by ATA abrogated anchorage-independent growth, the phenotype most closely linked to dysregulated TAZ/TEAD activity. Therefore, this study demonstrates that ATA is a novel small molecule that has the ability to disrupt the undruggable TAZ-TEAD interface.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

