Acute and persistent effects of commonly used antibiotics on the gut microbiome and resistome in healthy adults
- PMID: 35417701
- PMCID: PMC9066705
- DOI: 10.1016/j.celrep.2022.110649
Acute and persistent effects of commonly used antibiotics on the gut microbiome and resistome in healthy adults
Abstract
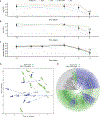
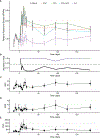
Antibiotics are deployed against bacterial pathogens, but their targeting of conserved microbial processes means they also collaterally perturb the commensal microbiome. To understand acute and persistent effects of antibiotics on the gut microbiota of healthy adult volunteers, we quantify microbiome dynamics before, during, and 6 months after exposure to 4 commonly used antibiotic regimens. We observe an acute decrease in species richness and culturable bacteria after antibiotics, with most healthy adult microbiomes returning to pre-treatment species richness after 2 months, but with an altered taxonomy, resistome, and metabolic output, as well as an increased antibiotic resistance burden. Azithromycin delays the recovery of species richness, resulting in greater compositional distance. A subset of volunteers experience a persistent reduction in microbiome diversity after antibiotics and share compositional similarities with patients hospitalized in intensive care units. These results improve our quantitative understanding of the impact of antibiotics on commensal microbiome dynamics, resilience, and recovery.
Keywords: CP: Microbiology; antibiotic resistance; host-microbe dynamics; microbial pathogenesis; microbiome; population dynamics; therapeutics.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Andremont A, Cervesi J, Bandinelli P-A, Vitry F, and De Gunzburg J (2021). Spare and repair the gut microbiota from antibiotic-induced dysbiosis: state-of-the-art. Drug Discov. Today 26, 2159–2163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

