HIV-1 Vpr drives a tissue residency-like phenotype during selective infection of resting memory T cells
- PMID: 35417711
- PMCID: PMC9350556
- DOI: 10.1016/j.celrep.2022.110650
HIV-1 Vpr drives a tissue residency-like phenotype during selective infection of resting memory T cells
Abstract
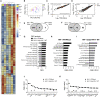
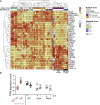
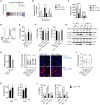
HIV-1 replicates in CD4+ T cells, leading to AIDS. Determining how HIV-1 shapes its niche to create a permissive environment is central to informing efforts to limit pathogenesis, disturb reservoirs, and achieve a cure. A key roadblock in understanding HIV-T cell interactions is the requirement to activate T cells in vitro to make them permissive to infection. This dramatically alters T cell biology and virus-host interactions. Here we show that HIV-1 cell-to-cell spread permits efficient, productive infection of resting memory T cells without prior activation. Strikingly, we find that HIV-1 infection primes resting T cells to gain characteristics of tissue-resident memory T cells (TRM), including upregulating key surface markers and the transcription factor Blimp-1 and inducing a transcriptional program overlapping the core TRM transcriptional signature. This reprogramming is driven by Vpr and requires Vpr packaging into virions and manipulation of STAT5. Thus, HIV-1 reprograms resting T cells, with implications for viral replication and persistence.
Keywords: CP: Immunology; CP: Microbiology; HIV-1; Vpr; cell-cell; permissivity; resting memory T cell; tissue residency; transcriptional reprogramming.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests L.J.P. participates in advisory boards and provides consultancy to SQZ Biotech.
Figures





References
-
- Agosto L.M., Herring M.B., Mothes W., Henderson A.J. HIV-1-Infected CD4+ T cells facilitate latent infection of resting CD4+ T cells through cell-cell contact. Cell Rep. 2018;24:2088–2100. - PubMed
-
- Almeida G.P. De, Lichtner P., Eckstein G., Brinkschmidt T., Chu C., Sun S., Reinhard J., Mädler S.C., Kloeppel M., Verbeek M., et al. Human skin-resident host T cells can persist long term after allogeneic stem cell transplantation and maintain recirculation potential. Sci. Immunol. 2022;7:eabe2634. - PubMed
-
- Amezcua Vesely M.C., Pallis P., Bielecki P., Low J.S., Zhao J., Harman C.C.D., Kroehling L., Jackson R., Bailis W., Licona-Limón P., et al. Effector TH17 cells give rise to long-lived TRM cells that are essential for an immediate response against bacterial infection. Cell. 2019;178:1176–1188.e15. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

