The tethered peptide activation mechanism of adhesion GPCRs
- PMID: 35418682
- PMCID: PMC9841879
- DOI: 10.1038/s41586-022-04575-7
The tethered peptide activation mechanism of adhesion GPCRs
Abstract
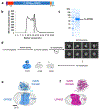
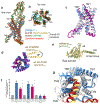
Adhesion G-protein-coupled receptors (aGPCRs) are characterized by the presence of auto-proteolysing extracellular regions that are involved in cell-cell and cell-extracellular matrix interactions1. Self cleavage within the aGPCR auto-proteolysis-inducing (GAIN) domain produces two protomers-N-terminal and C-terminal fragments-that remain non-covalently attached after receptors reach the cell surface1. Upon dissociation of the N-terminal fragment, the C-terminus of the GAIN domain acts as a tethered agonist (TA) peptide to activate the seven-transmembrane domain with a mechanism that has been poorly understood2-5. Here we provide cryo-electron microscopy snapshots of two distinct members of the aGPCR family, GPR56 (also known as ADGRG1) and latrophilin 3 (LPHN3 (also known as ADGRL3)). Low-resolution maps of the receptors in their N-terminal fragment-bound state indicate that the GAIN domain projects flexibly towards the extracellular space, keeping the encrypted TA peptide away from the seven-transmembrane domain. High-resolution structures of GPR56 and LPHN3 in their active, G-protein-coupled states, reveal that after dissociation of the extracellular region, the decrypted TA peptides engage the seven-transmembrane domain core with a notable conservation of interactions that also involve extracellular loop 2. TA binding stabilizes breaks in the middle of transmembrane helices 6 and 7 that facilitate aGPCR coupling and activation of heterotrimeric G proteins. Collectively, these results enable us to propose a general model for aGPCR activation.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures








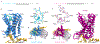
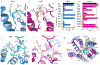
Comment in
-
Self-activated adhesion receptor proteins visualized.Nature. 2022 Apr;604(7907):628-630. doi: 10.1038/d41586-022-00972-0. Nature. 2022. PMID: 35418555 No abstract available.
-
Stachel-mediated activation of adhesion G protein-coupled receptors: insights from cryo-EM studies.Signal Transduct Target Ther. 2022 Jul 9;7(1):227. doi: 10.1038/s41392-022-01083-y. Signal Transduct Target Ther. 2022. PMID: 35810167 Free PMC article. No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

