Comprehensive assessment of cellular senescence in the tumor microenvironment
- PMID: 35419596
- PMCID: PMC9116224
- DOI: 10.1093/bib/bbac118
Comprehensive assessment of cellular senescence in the tumor microenvironment
Abstract
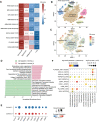
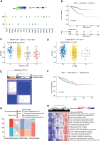
Cellular senescence (CS), a state of permanent growth arrest, is intertwined with tumorigenesis. Due to the absence of specific markers, characterizing senescence levels and senescence-related phenotypes across cancer types remain unexplored. Here, we defined computational metrics of senescence levels as CS scores to delineate CS landscape across 33 cancer types and 29 normal tissues and explored CS-associated phenotypes by integrating multiplatform data from ~20 000 patients and ~212 000 single-cell profiles. CS scores showed cancer type-specific associations with genomic and immune characteristics and significantly predicted immunotherapy responses and patient prognosis in multiple cancers. Single-cell CS quantification revealed intra-tumor heterogeneity and activated immune microenvironment in senescent prostate cancer. Using machine learning algorithms, we identified three CS genes as potential prognostic predictors in prostate cancer and verified them by immunohistochemical assays in 72 patients. Our study provides a comprehensive framework for evaluating senescence levels and clinical relevance, gaining insights into CS roles in cancer- and senescence-related biomarker discovery.
Keywords: cellular senescence; immunotherapy; machine learning; pan-cancer; single-cell.
© The Author(s) 2022. Published by Oxford University Press.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

