Ubiquitin ligase activity inhibits Cdk5 to control axon termination
- PMID: 35421092
- PMCID: PMC9041834
- DOI: 10.1371/journal.pgen.1010152
Ubiquitin ligase activity inhibits Cdk5 to control axon termination
Abstract
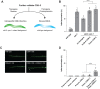
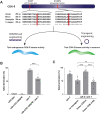
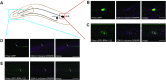
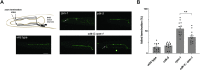
The Cdk5 kinase plays prominent roles in nervous system development, plasticity, behavior and disease. It also has important, non-neuronal functions in cancer, the immune system and insulin secretion. At present, we do not fully understand negative regulatory mechanisms that restrict Cdk5. Here, we use Caenorhabditis elegans to show that CDK-5 is inhibited by the RPM-1/FSN-1 ubiquitin ligase complex. This atypical RING ubiquitin ligase is conserved from C. elegans through mammals. Our finding originated from unbiased, in vivo affinity purification proteomics, which identified CDK-5 as a putative RPM-1 substrate. CRISPR-based, native biochemistry showed that CDK-5 interacts with the RPM-1/FSN-1 ubiquitin ligase complex. A CRISPR engineered RPM-1 substrate 'trap' enriched CDK-5 binding, which was mediated by the FSN-1 substrate recognition module. To test the functional genetic relationship between the RPM-1/FSN-1 ubiquitin ligase complex and CDK-5, we evaluated axon termination in mechanosensory neurons and motor neurons. Our results indicate that RPM-1/FSN-1 ubiquitin ligase activity restricts CDK-5 to control axon termination. Collectively, these proteomic, biochemical and genetic results increase our understanding of mechanisms that restrain Cdk5 in the nervous system.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

