Humanized mice reveal a macrophage-enriched gene signature defining human lung tissue protection during SARS-CoV-2 infection
- PMID: 35421379
- PMCID: PMC8977517
- DOI: 10.1016/j.celrep.2022.110714
Humanized mice reveal a macrophage-enriched gene signature defining human lung tissue protection during SARS-CoV-2 infection
Abstract
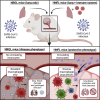
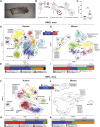
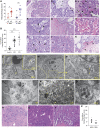
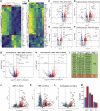
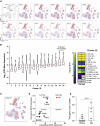
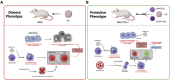
The human immunological mechanisms defining the clinical outcome of SARS-CoV-2 infection remain elusive. This knowledge gap is mostly driven by the lack of appropriate experimental platforms recapitulating human immune responses in a controlled human lung environment. Here, we report a mouse model (i.e., HNFL mice) co-engrafted with human fetal lung xenografts (fLX) and a myeloid-enhanced human immune system to identify cellular and molecular correlates of lung protection during SARS-CoV-2 infection. Unlike mice solely engrafted with human fLX, HNFL mice are protected against infection, severe inflammation, and histopathological phenotypes. Lung tissue protection from infection and severe histopathology associates with macrophage infiltration and differentiation and the upregulation of a macrophage-enriched signature composed of 11 specific genes mainly associated with the type I interferon signaling pathway. Our work highlights the HNFL model as a transformative platform to investigate, in controlled experimental settings, human myeloid immune mechanisms governing lung tissue protection during SARS-CoV-2 infection.
Keywords: COVID-19; CP: Immunology; CP: Microbiology; antiviral responses; human immune responses to SARS-CoV-2; humanized mice; macrophage responses to SARS-CoV-2; mouse models of SARS-CoV-2; pulmonary immune responses.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests K.P.F. is an employee of PerkinElmer, Inc., and a technology advisor of InVivo Analytics. N.P. and A.K. are shareholders of InVivo Analytics with issued patents.
Figures


References
-
- Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Publication types
MeSH terms
Grants and funding
- R01 HL139641/HL/NHLBI NIH HHS/United States
- S10 OD026983/OD/NIH HHS/United States
- R01 AI138797/AI/NIAID NIH HHS/United States
- R21 ES032882/ES/NIEHS NIH HHS/United States
- S10 OD030269/OD/NIH HHS/United States
- K22 AI144050/AI/NIAID NIH HHS/United States
- DP2 DA040254/DA/NIDA NIH HHS/United States
- R21 AI135517/AI/NIAID NIH HHS/United States
- R01 HL141513/HL/NHLBI NIH HHS/United States
- R01 AI107301/AI/NIAID NIH HHS/United States
- R01 AI153236/AI/NIAID NIH HHS/United States
- R01 AI153613/AI/NIAID NIH HHS/United States
- UL1 TR003017/TR/NCATS NIH HHS/United States
- R01 LM013154/LM/NLM NIH HHS/United States
- R01 AI146917/AI/NIAID NIH HHS/United States
- UL1 TR001430/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

