Validation of a small molecule inhibitor of PDE6D-RAS interaction with favorable anti-leukemic effects
- PMID: 35422065
- PMCID: PMC9010429
- DOI: 10.1038/s41408-022-00663-z
Validation of a small molecule inhibitor of PDE6D-RAS interaction with favorable anti-leukemic effects
Abstract
RAS mutations prevalent in high-risk leukemia have been linked to relapse and chemotherapy resistance. Efforts to directly target RAS proteins have been largely unsuccessful. However, since RAS-mediated transformation is dependent on signaling through the RAS-related C3 botulinum toxin substrate (RAC) small GTPase, we hypothesized that targeting RAC may be an effective therapeutic approach in RAS mutated tumors. Here we describe multiple small molecules capable of inhibiting RAC activation in acute lymphoblastic leukemia cell lines. One of these, DW0254, also demonstrates promising anti-leukemic activity in RAS-mutated cells. Using chemical proteomics and biophysical methods, we identified the hydrophobic pocket of phosphodiester 6 subunit delta (PDE6D), a known RAS chaperone, as a target for this compound. Inhibition of RAS localization to the plasma membrane upon DW0254 treatment is associated with RAC inhibition through a phosphatidylinositol-3-kinase/AKT-dependent mechanism. Our findings provide new insights into the importance of PDE6D-mediated transport for RAS-dependent RAC activation and leukemic cell survival.
© 2022. The Author(s).
Conflict of interest statement
DAW has been funded by the NIH. He is or was recently a member on a Board of Directors or advisory committees for: Bluebird bio, Orchard Therapeutics, Novartis, Beam Therapeutics, Emerging Therapy Solutions, Geneception, and BioMarin. In addition, he is the Co-founder of Alerion Biosciences and Orchard Therapeutics. AA, FA, EB, PK, and ME declare present or past employment by Evotec while engaged in the research project. SDV declares present employment by Novartis Institute for Biomedical Research. The remaining authors declare no competing interests.
Figures



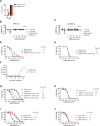

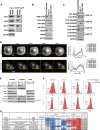
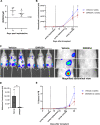
References
-
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–9. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

