Loss of MAT2A compromises methionine metabolism and represents a vulnerability in H3K27M mutant glioma by modulating the epigenome
- PMID: 35422502
- PMCID: PMC9551679
- DOI: 10.1038/s43018-022-00348-3
Loss of MAT2A compromises methionine metabolism and represents a vulnerability in H3K27M mutant glioma by modulating the epigenome
Erratum in
-
Author Correction: Loss of MAT2A compromises methionine metabolism and represents a vulnerability in H3K27M mutant glioma by modulating the epigenome.Nat Cancer. 2022 Jul;3(7):899. doi: 10.1038/s43018-022-00407-9. Nat Cancer. 2022. PMID: 35739422 No abstract available.
Abstract
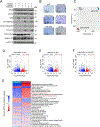
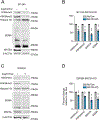
Diffuse midline gliomas (DMGs) bearing driver mutations of histone 3 lysine 27 (H3K27M) are incurable brain tumors with unique epigenomes. Here, we generated a syngeneic H3K27M mouse model to study the amino acid metabolic dependencies of these tumors. H3K27M mutant cells were highly dependent on methionine. Interrogating the methionine cycle dependency through a short-interfering RNA screen identified the enzyme methionine adenosyltransferase 2A (MAT2A) as a critical vulnerability in these tumors. This vulnerability was not mediated through the canonical mechanism of MTAP deletion; instead, DMG cells have lower levels of MAT2A protein, which is mediated by negative feedback induced by the metabolite decarboxylated S-adenosyl methionine. Depletion of residual MAT2A induces global depletion of H3K36me3, a chromatin mark of transcriptional elongation perturbing oncogenic and developmental transcriptional programs. Moreover, methionine-restricted diets extended survival in multiple models of DMG in vivo. Collectively, our results suggest that MAT2A presents an exploitable therapeutic vulnerability in H3K27M gliomas.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
















References
-
- Hoffman LM et al. Clinical, radiologic, pathologic, and molecular characteristics of long-term survivors of diffuse intrinsic pontine glioma (DIPG): a collaborative report from the International and European Society for Pediatric Oncology DIPG registries. J. Clin. Oncol 36, 1963–1972 (2018). - PMC - PubMed
-
- Sturm D et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

